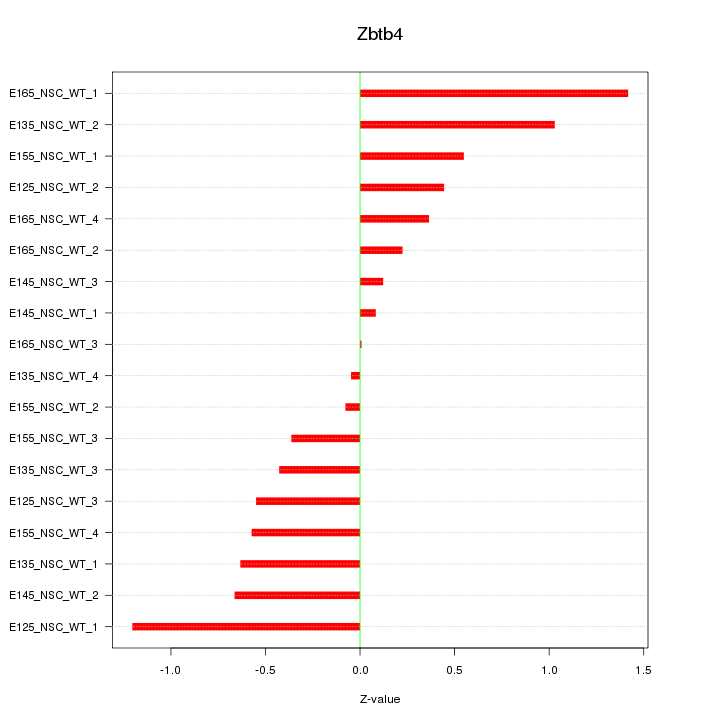
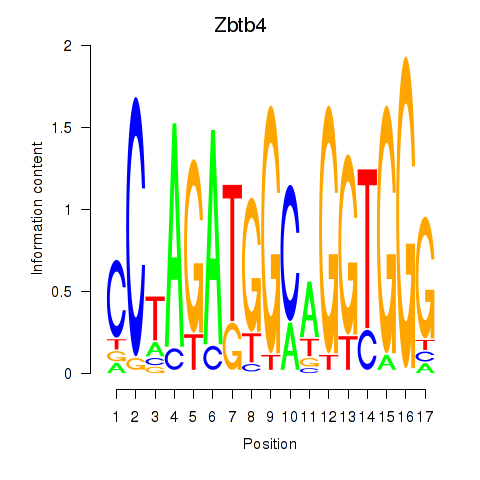
Motif ID: Zbtb4
Z-value: 0.625
Transcription factors associated with Zbtb4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Zbtb4 | ENSMUSG00000018750.8 | Zbtb4 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb4 | mm10_v2_chr11_+_69765970_69766027 | 0.55 | 1.9e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.2 | 0.9 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.2 | 0.8 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.2 | 0.6 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.2 | 0.6 | GO:1903002 | regulation of lipid transport across blood brain barrier(GO:1903000) positive regulation of lipid transport across blood brain barrier(GO:1903002) |
| 0.2 | 0.6 | GO:0032430 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) positive regulation of phospholipase A2 activity(GO:0032430) activation of meiosis involved in egg activation(GO:0060466) |
| 0.1 | 0.3 | GO:0003169 | coronary vein morphogenesis(GO:0003169) |
| 0.1 | 0.9 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 0.6 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.1 | 0.9 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 1.1 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.5 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.5 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 0.3 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 0.2 | GO:0021998 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) positive regulation of cardiac ventricle development(GO:1904414) |
| 0.1 | 0.2 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.1 | 0.2 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.1 | 0.5 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 0.6 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.4 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.1 | 0.3 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 0.6 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.6 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.3 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.1 | 0.4 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.4 | GO:0090298 | negative regulation of mitochondrial DNA replication(GO:0090298) |
| 0.1 | 1.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.2 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 0.6 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 0.5 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.1 | 0.2 | GO:0097494 | regulation of vesicle size(GO:0097494) |
| 0.0 | 0.3 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.3 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.2 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.0 | 0.2 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 1.1 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.1 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.2 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.7 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.6 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.1 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.4 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.0 | 0.3 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.5 | GO:0008210 | androgen metabolic process(GO:0008209) estrogen metabolic process(GO:0008210) |
| 0.0 | 0.3 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 | 0.4 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.3 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.1 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.0 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.1 | GO:0052428 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.5 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.0 | 0.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.0 | 0.1 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 0.0 | GO:1901079 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) positive regulation of relaxation of muscle(GO:1901079) |
| 0.0 | 0.1 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.0 | 0.1 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.0 | 0.1 | GO:0031034 | myosin filament assembly(GO:0031034) |
| 0.0 | 0.1 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.0 | 0.1 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.3 | GO:0014887 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.2 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.1 | GO:0033034 | positive regulation of neutrophil apoptotic process(GO:0033031) positive regulation of myeloid cell apoptotic process(GO:0033034) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 0.2 | 0.6 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 0.2 | 0.6 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.2 | 0.5 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 1.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 0.4 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 0.3 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.4 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.1 | 1.2 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.6 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.5 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.6 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.7 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.4 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.5 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 1.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.5 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.0 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.3 | 1.2 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.3 | 1.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 0.6 | GO:0046911 | hydroxyapatite binding(GO:0046848) metal chelating activity(GO:0046911) phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.2 | 0.7 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.1 | 1.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 0.9 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.7 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.1 | 0.6 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.1 | 0.6 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 0.3 | GO:0015173 | hydrogen:amino acid symporter activity(GO:0005280) aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.1 | 0.6 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.2 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 0.5 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.5 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 0.3 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 0.4 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.9 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.0 | 0.3 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.2 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.4 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.3 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.0 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.6 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.3 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.6 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.5 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.3 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 1.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | ST_WNT_CA2_CYCLIC_GMP_PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.0 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.4 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.0 | 0.7 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.6 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME_TRANSPORT_OF_ORGANIC_ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 1.9 | REACTOME_FATTY_ACYL_COA_BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.6 | REACTOME_HDL_MEDIATED_LIPID_TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.4 | REACTOME_COPI_MEDIATED_TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.6 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.9 | REACTOME_UNBLOCKING_OF_NMDA_RECEPTOR_GLUTAMATE_BINDING_AND_ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.4 | REACTOME_RESOLUTION_OF_AP_SITES_VIA_THE_SINGLE_NUCLEOTIDE_REPLACEMENT_PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.7 | REACTOME_INSULIN_SYNTHESIS_AND_PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.7 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.2 | REACTOME_ASSOCIATION_OF_LICENSING_FACTORS_WITH_THE_PRE_REPLICATIVE_COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.1 | REACTOME_TANDEM_PORE_DOMAIN_POTASSIUM_CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME_REGULATION_OF_RHEB_GTPASE_ACTIVITY_BY_AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.3 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.4 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.5 | REACTOME_SIGNALING_BY_ROBO_RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.6 | REACTOME_GLUCOSE_TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.1 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.3 | REACTOME_SYNTHESIS_OF_PC | Genes involved in Synthesis of PC |
| 0.0 | 0.3 | REACTOME_RAP1_SIGNALLING | Genes involved in Rap1 signalling |