Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
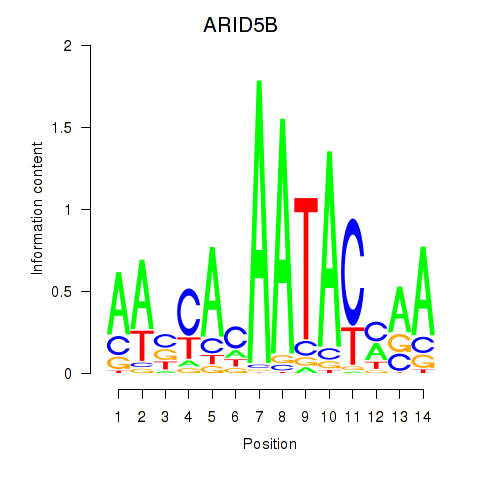
Results for ARID5B
Z-value: 0.18
Transcription factors associated with ARID5B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ARID5B
|
ENSG00000150347.10 | AT-rich interaction domain 5B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARID5B | hg19_v2_chr10_+_63661053_63661079 | 0.05 | 4.3e-01 | Click! |
Activity profile of ARID5B motif
Sorted Z-values of ARID5B motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_22090064 | 6.87 |
ENST00000339468.3
|
YPEL1
|
yippee-like 1 (Drosophila) |
| chr3_-_98241760 | 6.35 |
ENST00000507874.1
ENST00000502299.1 ENST00000508659.1 ENST00000510545.1 ENST00000511667.1 ENST00000394185.2 ENST00000394181.2 ENST00000508902.1 ENST00000341181.6 ENST00000437922.1 ENST00000394180.2 |
CLDND1
|
claudin domain containing 1 |
| chr3_-_98241358 | 6.03 |
ENST00000503004.1
ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1
|
claudin domain containing 1 |
| chr12_+_79258547 | 5.67 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr12_+_79258444 | 5.62 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr4_-_57524061 | 4.97 |
ENST00000508121.1
|
HOPX
|
HOP homeobox |
| chr22_-_22090043 | 4.71 |
ENST00000403503.1
|
YPEL1
|
yippee-like 1 (Drosophila) |
| chr8_-_101322132 | 4.55 |
ENST00000523481.1
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr5_-_88179302 | 4.42 |
ENST00000504921.2
|
MEF2C
|
myocyte enhancer factor 2C |
| chr17_-_3599696 | 4.22 |
ENST00000225328.5
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr7_-_37488834 | 4.05 |
ENST00000310758.4
|
ELMO1
|
engulfment and cell motility 1 |
| chr4_-_71532339 | 3.88 |
ENST00000254801.4
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr17_-_3599492 | 3.85 |
ENST00000435558.1
ENST00000345901.3 |
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr18_-_53303123 | 3.84 |
ENST00000569357.1
ENST00000565124.1 ENST00000398339.1 |
TCF4
|
transcription factor 4 |
| chr1_+_151739131 | 3.48 |
ENST00000400999.1
|
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr4_+_76995855 | 3.45 |
ENST00000355810.4
ENST00000349321.3 |
ART3
|
ADP-ribosyltransferase 3 |
| chr18_+_32556892 | 3.30 |
ENST00000591734.1
ENST00000413393.1 ENST00000589180.1 ENST00000587359.1 |
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr11_-_111794446 | 3.22 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr8_-_101321584 | 3.20 |
ENST00000523167.1
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr3_+_43328004 | 3.12 |
ENST00000454177.1
ENST00000429705.2 ENST00000296088.7 ENST00000437827.1 |
SNRK
|
SNF related kinase |
| chr5_-_88179017 | 2.98 |
ENST00000514028.1
ENST00000514015.1 ENST00000503075.1 ENST00000437473.2 |
MEF2C
|
myocyte enhancer factor 2C |
| chr7_+_153749732 | 2.97 |
ENST00000377770.3
|
DPP6
|
dipeptidyl-peptidase 6 |
| chr17_-_76713100 | 2.96 |
ENST00000585509.1
|
CYTH1
|
cytohesin 1 |
| chr7_-_104909435 | 2.95 |
ENST00000357311.3
|
SRPK2
|
SRSF protein kinase 2 |
| chr1_+_158975744 | 2.83 |
ENST00000426592.2
|
IFI16
|
interferon, gamma-inducible protein 16 |
| chr11_-_73882029 | 2.66 |
ENST00000539061.1
|
C2CD3
|
C2 calcium-dependent domain containing 3 |
| chr15_+_25068773 | 2.61 |
ENST00000400100.1
ENST00000400098.1 |
SNRPN
|
small nuclear ribonucleoprotein polypeptide N |
| chrX_+_128913906 | 2.51 |
ENST00000356892.3
|
SASH3
|
SAM and SH3 domain containing 3 |
| chr7_-_56119156 | 2.51 |
ENST00000421312.1
ENST00000416592.1 |
PSPH
|
phosphoserine phosphatase |
| chr10_+_126150369 | 2.48 |
ENST00000392757.4
ENST00000368842.5 ENST00000368839.1 |
LHPP
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr13_+_52436111 | 2.28 |
ENST00000242819.4
|
CCDC70
|
coiled-coil domain containing 70 |
| chr2_-_158345462 | 2.05 |
ENST00000439355.1
ENST00000540637.1 |
CYTIP
|
cytohesin 1 interacting protein |
| chr2_-_170430277 | 1.93 |
ENST00000438035.1
ENST00000453929.2 |
FASTKD1
|
FAST kinase domains 1 |
| chr15_-_80263506 | 1.93 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr2_-_170430366 | 1.83 |
ENST00000453153.2
ENST00000445210.1 |
FASTKD1
|
FAST kinase domains 1 |
| chrX_-_140786896 | 1.80 |
ENST00000370515.3
|
SPANXD
|
SPANX family, member D |
| chr19_+_56459198 | 1.77 |
ENST00000291971.3
ENST00000590542.1 |
NLRP8
|
NLR family, pyrin domain containing 8 |
| chr1_-_153085984 | 1.71 |
ENST00000468739.1
|
SPRR2F
|
small proline-rich protein 2F |
| chr9_+_114287433 | 1.64 |
ENST00000358151.4
ENST00000355824.3 ENST00000374374.3 ENST00000309235.5 |
ZNF483
|
zinc finger protein 483 |
| chr7_-_24797546 | 1.63 |
ENST00000414428.1
ENST00000419307.1 ENST00000342947.3 |
DFNA5
|
deafness, autosomal dominant 5 |
| chr12_+_56324756 | 1.60 |
ENST00000331886.5
ENST00000555090.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr17_+_3379284 | 1.58 |
ENST00000263080.2
|
ASPA
|
aspartoacylase |
| chr3_-_52713729 | 1.56 |
ENST00000296302.7
ENST00000356770.4 ENST00000337303.4 ENST00000409057.1 ENST00000410007.1 ENST00000409114.3 ENST00000409767.1 ENST00000423351.1 |
PBRM1
|
polybromo 1 |
| chr5_+_173763250 | 1.56 |
ENST00000515513.1
ENST00000507361.1 ENST00000510234.1 |
RP11-267A15.1
|
RP11-267A15.1 |
| chrX_+_140677562 | 1.47 |
ENST00000370518.3
|
SPANXA2
|
SPANX family, member A2 |
| chr2_-_74618964 | 1.46 |
ENST00000417090.1
ENST00000409868.1 |
DCTN1
|
dynactin 1 |
| chr3_-_79816965 | 1.46 |
ENST00000464233.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr4_+_71588372 | 1.44 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr2_+_89999259 | 1.43 |
ENST00000558026.1
|
IGKV2D-28
|
immunoglobulin kappa variable 2D-28 |
| chr17_-_10452929 | 1.42 |
ENST00000532183.2
ENST00000397183.2 ENST00000420805.1 |
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult |
| chr7_+_116654935 | 1.41 |
ENST00000432298.1
ENST00000422922.1 |
ST7
|
suppression of tumorigenicity 7 |
| chr12_+_56324933 | 1.36 |
ENST00000549629.1
ENST00000555218.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr7_+_149535455 | 1.33 |
ENST00000223210.4
ENST00000460379.1 |
ZNF862
|
zinc finger protein 862 |
| chr2_-_74619152 | 1.29 |
ENST00000440727.1
ENST00000409240.1 |
DCTN1
|
dynactin 1 |
| chr1_+_104159999 | 1.26 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr20_-_9819674 | 1.26 |
ENST00000378429.3
|
PAK7
|
p21 protein (Cdc42/Rac)-activated kinase 7 |
| chr1_+_43855560 | 1.18 |
ENST00000562955.1
|
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr5_+_446253 | 1.16 |
ENST00000315013.5
|
EXOC3
|
exocyst complex component 3 |
| chr1_+_78245303 | 1.16 |
ENST00000370791.3
ENST00000443751.2 |
FAM73A
|
family with sequence similarity 73, member A |
| chr2_-_89327228 | 1.15 |
ENST00000483158.1
|
IGKV3-11
|
immunoglobulin kappa variable 3-11 |
| chr12_-_118796910 | 1.15 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr1_-_153066998 | 1.11 |
ENST00000368750.3
|
SPRR2E
|
small proline-rich protein 2E |
| chrX_-_140336629 | 1.11 |
ENST00000358993.2
|
SPANXC
|
SPANX family, member C |
| chr6_+_32812568 | 1.07 |
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr11_-_74178582 | 1.05 |
ENST00000531854.1
ENST00000526855.1 ENST00000529425.1 ENST00000310128.4 ENST00000532569.1 |
KCNE3
|
potassium voltage-gated channel, Isk-related family, member 3 |
| chr1_+_92632542 | 1.04 |
ENST00000409154.4
ENST00000370378.4 |
KIAA1107
|
KIAA1107 |
| chrX_+_140084756 | 1.03 |
ENST00000449283.1
|
SPANXB2
|
SPANX family, member B2 |
| chr1_-_114414316 | 1.03 |
ENST00000528414.1
ENST00000538253.1 ENST00000460620.1 ENST00000420377.2 ENST00000525799.1 ENST00000359785.5 |
PTPN22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr15_-_91565743 | 0.98 |
ENST00000535843.1
|
VPS33B
|
vacuolar protein sorting 33 homolog B (yeast) |
| chr14_-_24711865 | 0.97 |
ENST00000399423.4
ENST00000267415.7 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr5_+_118691706 | 0.97 |
ENST00000415806.2
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr5_+_131993856 | 0.96 |
ENST00000304506.3
|
IL13
|
interleukin 13 |
| chr6_-_160210715 | 0.94 |
ENST00000392168.2
ENST00000321394.7 |
TCP1
|
t-complex 1 |
| chrX_-_33229636 | 0.92 |
ENST00000357033.4
|
DMD
|
dystrophin |
| chr13_-_41706864 | 0.91 |
ENST00000379485.1
ENST00000499385.2 |
KBTBD6
|
kelch repeat and BTB (POZ) domain containing 6 |
| chr1_-_144994909 | 0.90 |
ENST00000369347.4
ENST00000369354.3 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr15_-_91565770 | 0.86 |
ENST00000535906.1
ENST00000333371.3 |
VPS33B
|
vacuolar protein sorting 33 homolog B (yeast) |
| chr4_+_106631966 | 0.85 |
ENST00000360505.5
ENST00000510865.1 ENST00000509336.1 |
GSTCD
|
glutathione S-transferase, C-terminal domain containing |
| chr4_-_90229142 | 0.84 |
ENST00000609438.1
|
GPRIN3
|
GPRIN family member 3 |
| chr14_-_24711806 | 0.84 |
ENST00000540705.1
ENST00000538777.1 ENST00000558566.1 ENST00000559019.1 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr14_+_64680854 | 0.84 |
ENST00000458046.2
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2 |
| chr19_+_17326141 | 0.77 |
ENST00000445667.2
ENST00000263897.5 |
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr1_-_76076759 | 0.75 |
ENST00000370855.5
|
SLC44A5
|
solute carrier family 44, member 5 |
| chr5_+_92919043 | 0.72 |
ENST00000327111.3
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr1_+_32608566 | 0.69 |
ENST00000545542.1
|
KPNA6
|
karyopherin alpha 6 (importin alpha 7) |
| chr22_+_40573921 | 0.68 |
ENST00000454349.2
ENST00000335727.9 |
TNRC6B
|
trinucleotide repeat containing 6B |
| chr3_+_32726774 | 0.68 |
ENST00000538368.1
|
CNOT10
|
CCR4-NOT transcription complex, subunit 10 |
| chr1_-_144995074 | 0.68 |
ENST00000534536.1
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr5_+_161495038 | 0.65 |
ENST00000393933.4
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr19_-_56826157 | 0.61 |
ENST00000592509.1
ENST00000592679.1 ENST00000588442.1 ENST00000593106.1 ENST00000587492.1 ENST00000254165.3 |
ZSCAN5A
|
zinc finger and SCAN domain containing 5A |
| chrX_-_57164058 | 0.61 |
ENST00000374906.3
|
SPIN2A
|
spindlin family, member 2A |
| chr1_-_144994840 | 0.60 |
ENST00000369351.3
ENST00000369349.3 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr22_+_19467261 | 0.60 |
ENST00000455750.1
ENST00000437685.2 ENST00000263201.1 ENST00000404724.3 |
CDC45
|
cell division cycle 45 |
| chr22_+_39916558 | 0.56 |
ENST00000337304.2
ENST00000396680.1 |
ATF4
|
activating transcription factor 4 |
| chrX_+_54466829 | 0.56 |
ENST00000375151.4
|
TSR2
|
TSR2, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr6_-_30080876 | 0.55 |
ENST00000376734.3
|
TRIM31
|
tripartite motif containing 31 |
| chr4_-_80247162 | 0.55 |
ENST00000286794.4
|
NAA11
|
N(alpha)-acetyltransferase 11, NatA catalytic subunit |
| chr8_-_145652336 | 0.51 |
ENST00000529182.1
ENST00000526054.1 |
VPS28
|
vacuolar protein sorting 28 homolog (S. cerevisiae) |
| chr17_-_36358166 | 0.50 |
ENST00000537432.1
|
TBC1D3
|
TBC1 domain family, member 3 |
| chr4_-_89978299 | 0.48 |
ENST00000511976.1
ENST00000509094.1 ENST00000264344.5 ENST00000515600.1 |
FAM13A
|
family with sequence similarity 13, member A |
| chr18_-_25616519 | 0.47 |
ENST00000399380.3
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr12_+_120933859 | 0.47 |
ENST00000242577.6
ENST00000548214.1 ENST00000392508.2 |
DYNLL1
|
dynein, light chain, LC8-type 1 |
| chr12_+_120933904 | 0.46 |
ENST00000550178.1
ENST00000550845.1 ENST00000549989.1 ENST00000552870.1 |
DYNLL1
|
dynein, light chain, LC8-type 1 |
| chr8_-_66474884 | 0.45 |
ENST00000520902.1
|
CTD-3025N20.2
|
CTD-3025N20.2 |
| chr12_+_57146233 | 0.45 |
ENST00000554643.1
ENST00000556650.1 ENST00000554150.1 ENST00000554155.1 |
HSD17B6
|
hydroxysteroid (17-beta) dehydrogenase 6 |
| chr1_-_115292591 | 0.44 |
ENST00000438362.2
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr12_-_10007448 | 0.40 |
ENST00000538152.1
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr11_-_78052923 | 0.39 |
ENST00000340149.2
|
GAB2
|
GRB2-associated binding protein 2 |
| chr1_+_3773825 | 0.35 |
ENST00000378209.3
ENST00000338895.3 ENST00000378212.2 ENST00000341385.3 |
DFFB
|
DNA fragmentation factor, 40kDa, beta polypeptide (caspase-activated DNase) |
| chr5_-_64920115 | 0.33 |
ENST00000381018.3
ENST00000274327.7 |
TRIM23
|
tripartite motif containing 23 |
| chr18_+_32173276 | 0.32 |
ENST00000591816.1
ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA
|
dystrobrevin, alpha |
| chr11_-_33774944 | 0.31 |
ENST00000532057.1
ENST00000531080.1 |
FBXO3
|
F-box protein 3 |
| chr1_-_168698433 | 0.30 |
ENST00000367817.3
|
DPT
|
dermatopontin |
| chr1_+_104293028 | 0.30 |
ENST00000370079.3
|
AMY1C
|
amylase, alpha 1C (salivary) |
| chr5_+_137225158 | 0.29 |
ENST00000290431.5
|
PKD2L2
|
polycystic kidney disease 2-like 2 |
| chr6_+_25754927 | 0.28 |
ENST00000377905.4
ENST00000439485.2 |
SLC17A4
|
solute carrier family 17, member 4 |
| chr9_+_706842 | 0.26 |
ENST00000382293.3
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr9_+_116267536 | 0.25 |
ENST00000374136.1
|
RGS3
|
regulator of G-protein signaling 3 |
| chr1_+_152975488 | 0.23 |
ENST00000542696.1
|
SPRR3
|
small proline-rich protein 3 |
| chr11_-_102714534 | 0.22 |
ENST00000299855.5
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr1_+_44435646 | 0.20 |
ENST00000255108.3
ENST00000412950.2 ENST00000396758.2 |
DPH2
|
DPH2 homolog (S. cerevisiae) |
| chr6_+_122720681 | 0.19 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chr7_-_56119238 | 0.19 |
ENST00000275605.3
ENST00000395471.3 |
PSPH
|
phosphoserine phosphatase |
| chr1_+_150039369 | 0.18 |
ENST00000369130.3
ENST00000369128.5 |
VPS45
|
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr7_-_155604967 | 0.16 |
ENST00000297261.2
|
SHH
|
sonic hedgehog |
| chr5_+_135496675 | 0.15 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chr11_+_73882144 | 0.14 |
ENST00000328257.8
|
PPME1
|
protein phosphatase methylesterase 1 |
| chr2_+_223725652 | 0.14 |
ENST00000357430.3
ENST00000392066.3 |
ACSL3
|
acyl-CoA synthetase long-chain family member 3 |
| chr22_-_42086477 | 0.14 |
ENST00000402458.1
|
NHP2L1
|
NHP2 non-histone chromosome protein 2-like 1 (S. cerevisiae) |
| chr4_-_159094194 | 0.14 |
ENST00000592057.1
ENST00000585682.1 ENST00000393807.5 |
FAM198B
|
family with sequence similarity 198, member B |
| chr7_-_115608304 | 0.13 |
ENST00000457268.1
|
TFEC
|
transcription factor EC |
| chr10_+_53806501 | 0.13 |
ENST00000373975.2
|
PRKG1
|
protein kinase, cGMP-dependent, type I |
| chr18_+_21032781 | 0.13 |
ENST00000339486.3
|
RIOK3
|
RIO kinase 3 |
| chr6_-_30080863 | 0.11 |
ENST00000540829.1
|
TRIM31
|
tripartite motif containing 31 |
| chr14_+_22458631 | 0.11 |
ENST00000390444.1
|
TRAV16
|
T cell receptor alpha variable 16 |
| chr1_-_31538517 | 0.10 |
ENST00000440538.2
ENST00000423018.2 ENST00000424085.2 ENST00000426105.2 ENST00000257075.5 ENST00000373747.3 ENST00000525843.1 ENST00000373742.2 |
PUM1
|
pumilio RNA-binding family member 1 |
| chr2_+_181845843 | 0.09 |
ENST00000602710.1
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chrX_-_11308598 | 0.06 |
ENST00000380717.3
|
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr5_-_179047881 | 0.03 |
ENST00000521173.1
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr12_-_67197760 | 0.03 |
ENST00000539540.1
ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr16_+_58426296 | 0.03 |
ENST00000426538.2
ENST00000328514.7 ENST00000318129.5 |
GINS3
|
GINS complex subunit 3 (Psf3 homolog) |
| chr5_+_175288631 | 0.02 |
ENST00000509837.1
|
CPLX2
|
complexin 2 |
| chr7_+_138145145 | 0.02 |
ENST00000415680.2
|
TRIM24
|
tripartite motif containing 24 |
| chr3_+_184018352 | 0.01 |
ENST00000435761.1
ENST00000439383.1 |
PSMD2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| chr9_+_125796806 | 0.00 |
ENST00000373642.1
|
GPR21
|
G protein-coupled receptor 21 |
| chr11_+_73882311 | 0.00 |
ENST00000398427.4
ENST00000544401.1 |
PPME1
|
protein phosphatase methylesterase 1 |
| chr2_-_178753465 | 0.00 |
ENST00000389683.3
|
PDE11A
|
phosphodiesterase 11A |
Network of associatons between targets according to the STRING database.
First level regulatory network of ARID5B
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.3 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 1.8 | 7.4 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.7 | 3.5 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.7 | 2.7 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.6 | 2.9 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.5 | 1.6 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.5 | 1.5 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.3 | 1.0 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.3 | 1.0 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.3 | 2.7 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.3 | 1.0 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.3 | 1.8 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.3 | 8.1 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.2 | 4.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.2 | 1.8 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.2 | 5.0 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.2 | 2.7 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.2 | 0.6 | GO:0031938 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) regulation of chromatin silencing at telomere(GO:0031938) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.2 | 1.2 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.2 | 0.6 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.2 | 1.3 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.2 | 0.5 | GO:2000395 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) ubiquitin-dependent endocytosis(GO:0070086) regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.1 | 3.0 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.8 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 0.5 | GO:0017198 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 | 3.0 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 1.4 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 2.5 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.1 | 0.4 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.1 | 1.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 4.0 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 3.1 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 0.5 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 | 0.9 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.3 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.9 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.7 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 1.9 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.4 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 7.8 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.1 | 2.8 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 0.2 | GO:2000729 | bud outgrowth involved in lung branching(GO:0060447) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 1.4 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.1 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 1.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.7 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.3 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 2.0 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.6 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.7 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.2 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 1.6 | GO:0060113 | inner ear receptor cell differentiation(GO:0060113) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.2 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 2.5 | GO:0009168 | purine nucleoside monophosphate biosynthetic process(GO:0009127) purine ribonucleoside monophosphate biosynthetic process(GO:0009168) |
| 0.0 | 0.4 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 1.8 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.0 | 0.7 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.7 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 3.1 | GO:0045333 | cellular respiration(GO:0045333) |
| 0.0 | 2.6 | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.0 | 0.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 1.1 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 1.5 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 2.8 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 11.3 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.5 | 1.4 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.4 | 8.1 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.4 | 1.8 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.3 | 4.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 1.8 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.2 | 3.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 0.6 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.2 | 1.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.2 | 6.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.2 | 2.6 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 1.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.8 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.9 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 2.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.6 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 0.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.9 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 1.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 3.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.5 | GO:0005916 | fascia adherens(GO:0005916) catenin complex(GO:0016342) |
| 0.0 | 0.4 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 1.6 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 1.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.7 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 6.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 12.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 7.3 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 2.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 3.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 2.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 10.5 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 4.0 | GO:0000790 | nuclear chromatin(GO:0000790) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 11.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.7 | 8.1 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.7 | 3.5 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.7 | 3.5 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.6 | 2.5 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.5 | 2.7 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.4 | 6.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.3 | 1.3 | GO:0016160 | amylase activity(GO:0016160) |
| 0.3 | 3.8 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 1.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 1.6 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.2 | 7.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.2 | 0.7 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.2 | 3.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.5 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.1 | 3.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 3.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 3.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.9 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 2.9 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 2.7 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 1.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.9 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.6 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.9 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.4 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.6 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 1.4 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 1.8 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 1.1 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.6 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 5.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 1.0 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.3 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 2.6 | GO:0000287 | magnesium ion binding(GO:0000287) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 11.3 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 7.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 1.0 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 4.0 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 3.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 2.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 3.0 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 3.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 2.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 11.3 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.2 | 11.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 2.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 2.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 4.0 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 3.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 4.6 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 0.5 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.9 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 3.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 1.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.7 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |