Mean activities
Top 10 and bottom 10 mean activities.
Mean activities sorted by motif Z-value.
| Motif name | Mean activity | Z-Value | Associated genes | Logo |
|---|---|---|---|---|
| AAAGUGC | 0.055 | 0.162 |
|

|
| AACACUG | -0.303 | -0.622 |
|

|
| AACAGUC | -0.166 | -0.210 |
|

|
| AACCGUU | -0.796 | -0.153 |
|
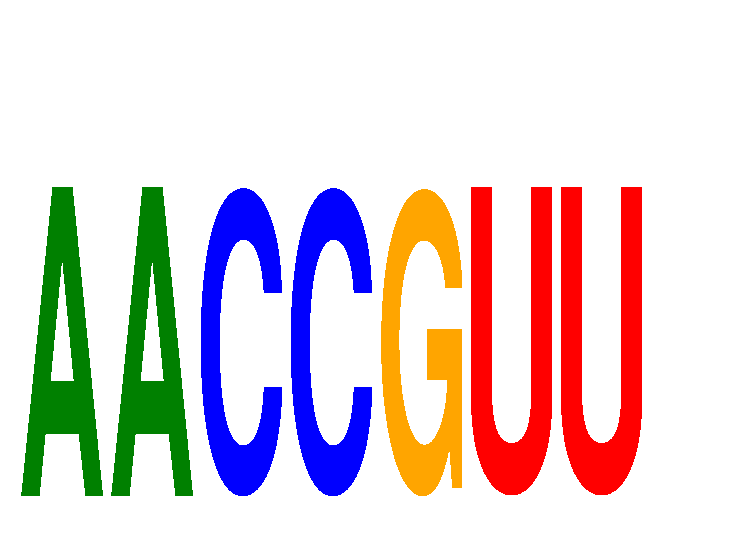
|
| AACCUGG | -0.279 | -0.105 |
|
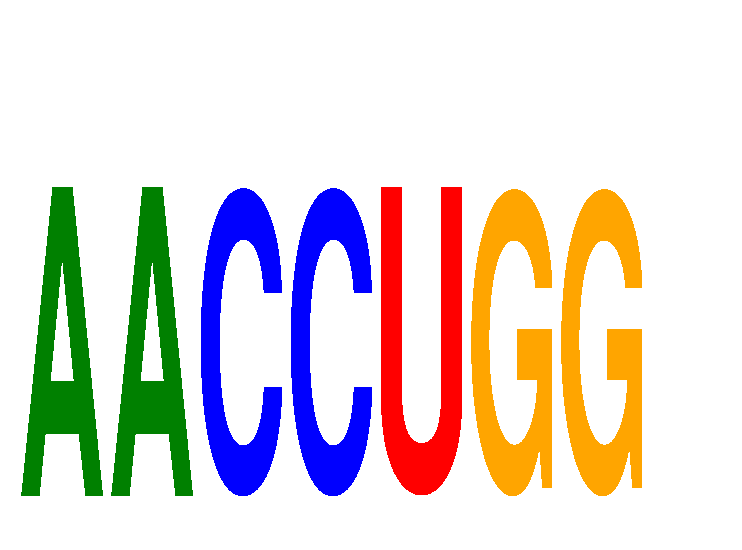
|
| AACGGAA | 0.367 | 0.156 |
|
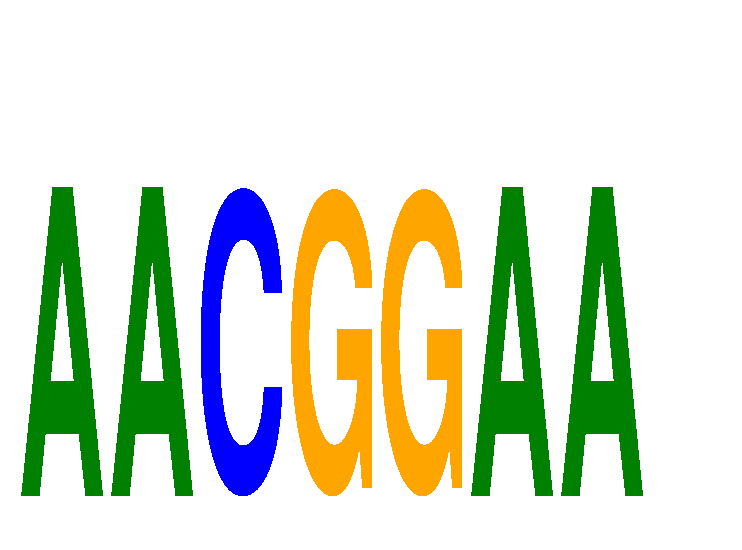
|
| AAGGCAC | 0.014 | 0.049 |
|

|
| AAGGUGC | -0.024 | -0.030 |
|
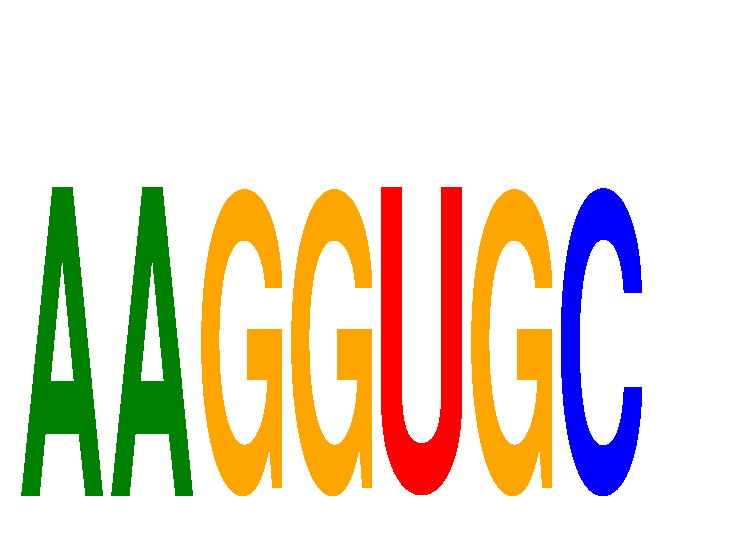
|
| AAGUGCU | -0.336 | -0.723 |
|

|
| AAUACUG | 0.052 | 0.156 |
|

|
| AAUCUCA | -0.037 | -0.024 |
|

|
| AAUCUCU | -0.065 | -0.038 |
|

|
| AAUGCCC | -0.058 | -0.067 |
|
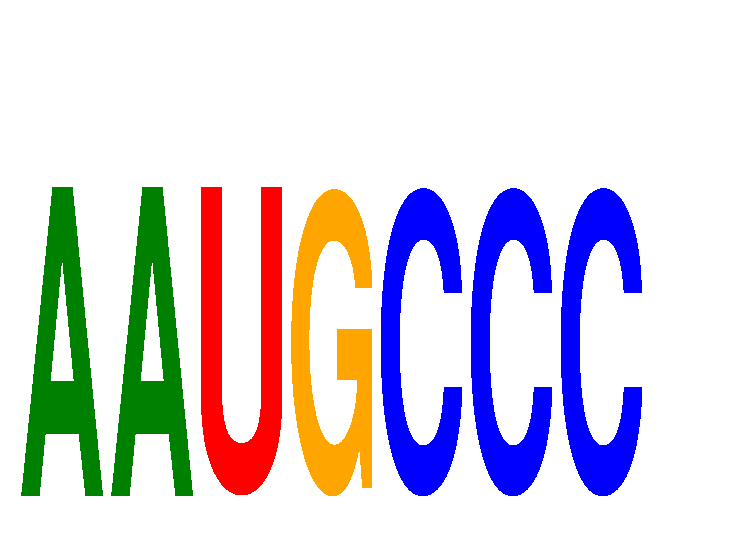
|
| ACAGUAC | -0.033 | -0.065 |
|

|
| ACAGUAU | -0.081 | -0.152 |
|

|
| ACAUUCA | 0.087 | 0.243 |
|

|
| ACCACAG | 0.298 | 0.226 |
|

|
| ACCCGUA | -1.508 | -0.359 |
|

|
| ACCCUGU | -0.010 | -0.012 |
|
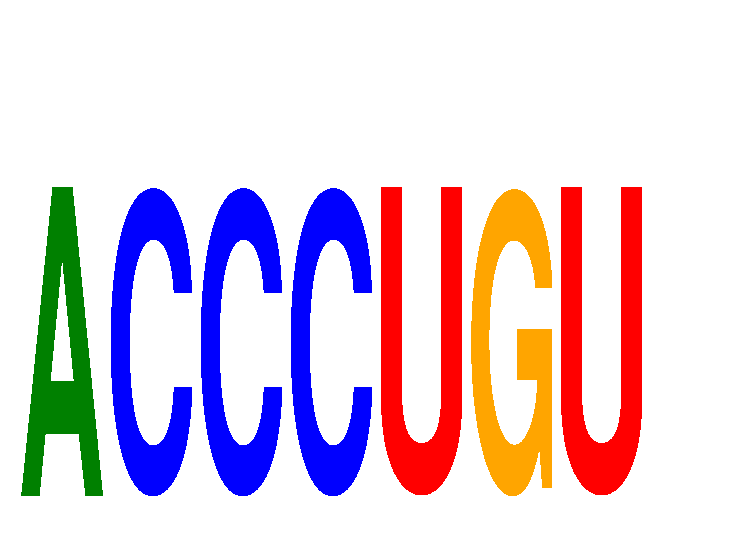
|
| ACUGCAU | 0.104 | 0.101 |
|

|
| ACUGGCC | -0.181 | -0.188 |
|

|
| AGAUCAG | -0.036 | -0.007 |
|

|
| AGCACCA | -0.071 | -0.256 |
|

|
| AGCAGCA | 0.108 | 0.290 |
|

|
| AGCAGCG | -0.160 | -0.282 |
|

|
| AGCCCUU | -0.192 | -0.334 |
|

|
| AGCUGCC | -0.113 | -0.223 |
|

|
| AGCUUAU | 0.444 | 0.557 |
|

|
| AGGUAGU | -0.239 | -0.393 |
|

|
| AGUGCAA | -0.028 | -0.073 |
|

|
| AGUGCUU | 0.070 | 0.145 |
|

|
| AGUGGUU | -0.480 | -0.729 |
|

|
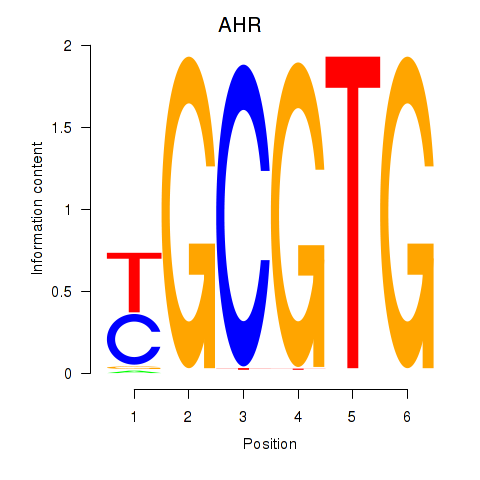
| AHR_ARNT2 | 0.039 | 0.399 |
|
|
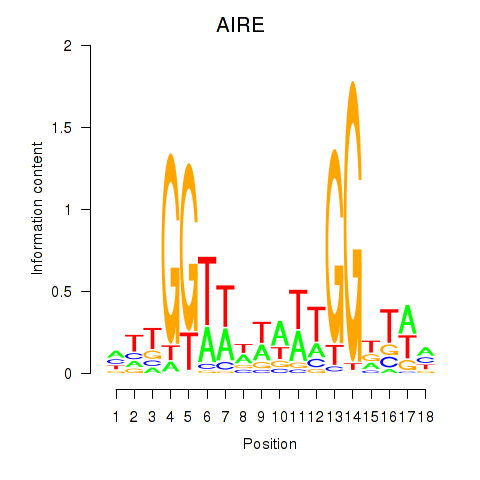
| AIRE | 0.084 | 0.152 |
|
|
| ALX1_ARX | -0.437 | -0.508 |
|
|
| ALX3 | 0.019 | 0.035 |
|
|
| ARID3A | 0.030 | 0.099 |
|

|
| ARID5A | 0.024 | 0.079 |
|
|
| ARID5B | 0.086 | 0.183 |
|
|
| ARNT | 0.215 | 0.733 |
|

|
| AR_NR3C2 | 0.064 | 0.119 |
|
|
| ATF2_ATF1_ATF3 | 0.083 | 0.552 |
|
|
| ATF4 | 0.237 | 0.394 |
|

|
| ATF5 | -0.067 | -0.155 |
|
|
| ATF6 | 0.196 | 0.666 |
|

|
| ATF7 | -0.215 | -0.520 |
|
|
| AUAAAGU | 0.268 | 0.438 |
|

|
| AUGACAC | 1.360 | 0.391 |
|

|
| AUGGCAC | 0.394 | 0.661 |
|

|
| AUGGCUU | -0.220 | -0.574 |
|

|
| AUGUGCC | 0.480 | 0.434 |
|

|
| AUUGCAC | -0.076 | -0.222 |
|

|
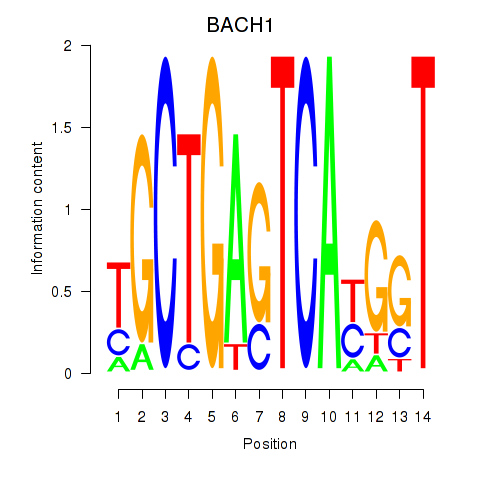
| BACH1_NFE2_NFE2L2 | 0.452 | 1.368 |
|
|
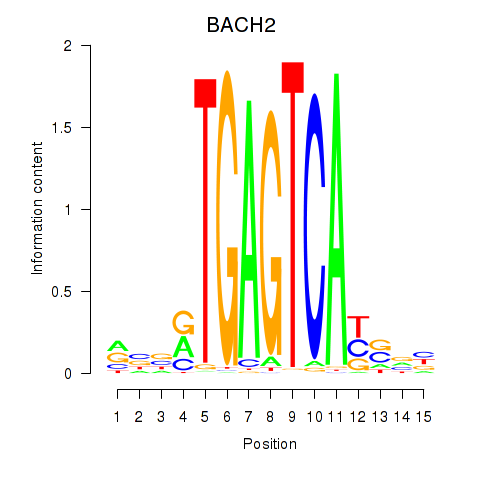
| BACH2 | 0.420 | 0.896 |
|
|
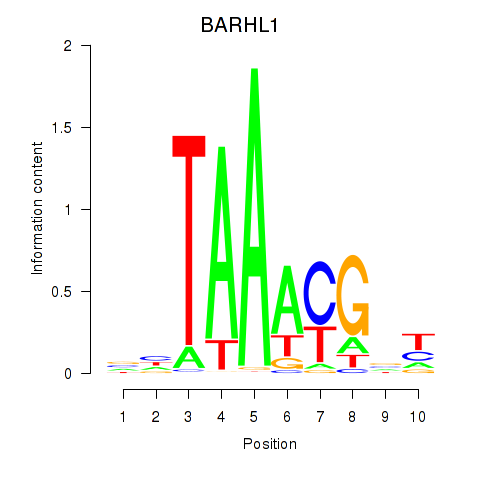
| BARHL1 | 0.030 | 0.251 |
|
|
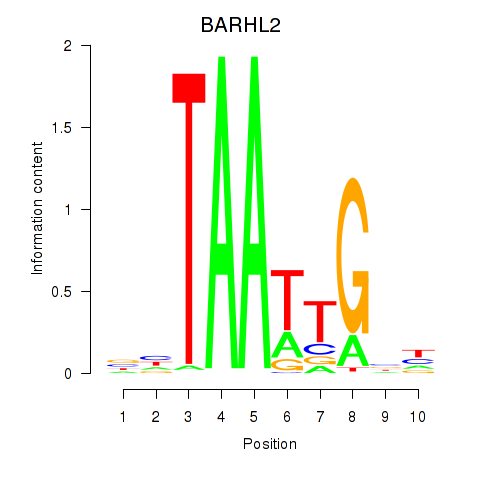
| BARHL2 | -0.081 | -0.294 |
|
|
| BARX1 | 0.139 | 0.604 |
|
|
| BATF | 0.140 | 1.016 |
|
|
| BATF3 | 0.004 | 0.006 |
|
|
| BBX | 0.044 | 0.064 |
|
|
| BCL6B | 0.417 | 0.810 |
|
|
| BHLHE22_BHLHA15_BHLHE23 | -0.091 | -0.140 |
|
|
| BHLHE40 | -0.288 | -0.569 |
|
|
| BPTF | -0.008 | -0.066 |
|
|
| BRCA1 | 0.193 | 0.251 |
|
|
| CACAGUG | 0.168 | 0.332 |
|

|
| CACCUCC | 3.170 | 0.846 |
|

|
| CAGUAGU | -0.364 | -0.677 |
|

|
| CAGUCCA | -0.033 | -0.029 |
|

|
| CAGUGCA | -0.132 | -0.305 |
|

|
| CBFB | 0.071 | 0.090 |
|
|
| CCACAGG | -0.077 | -0.057 |
|

|
| CCAGCAU | 0.080 | 0.056 |
|

|
| CCAGUGU | -0.090 | -0.188 |
|

|
| CCCUGAG | -0.347 | -0.971 |
|
|
| CCUUCAU | 0.537 | 0.591 |
|

|
| CCUUGGC | 0.590 | 0.247 |
|

|
| CDC5L | -0.038 | -0.151 |
|
|
| CDX1 | 0.037 | 0.110 |
|
|
| CDX2 | 0.429 | 0.810 |
|
|
| CEBPA | -0.169 | -0.610 |
|
|
| CEBPB | -0.022 | -0.052 |
|
|
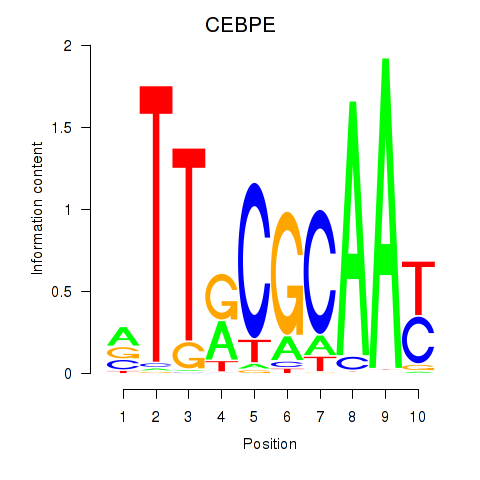
| CEBPE_CEBPD | 0.034 | 0.101 |
|
|
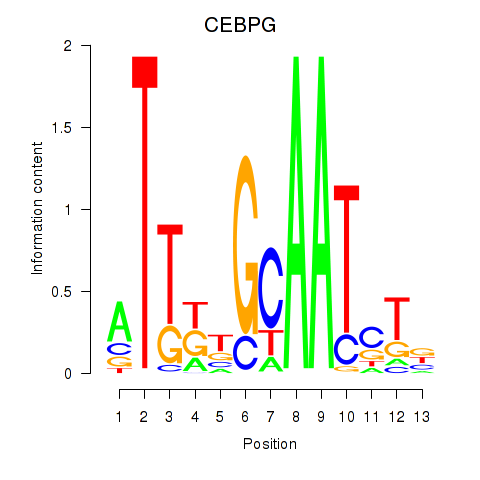
| CEBPG | 0.072 | 0.134 |
|
|
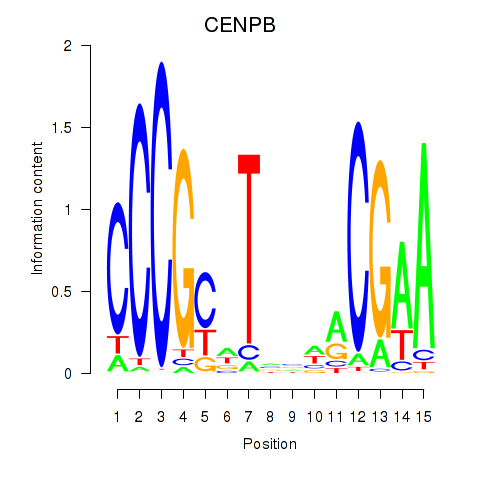
| CENPB | 0.137 | 0.265 |
|
|
| CGUACCG | -0.313 | -0.101 |
|

|
| CGUGUCU | -0.152 | -0.029 |
|

|
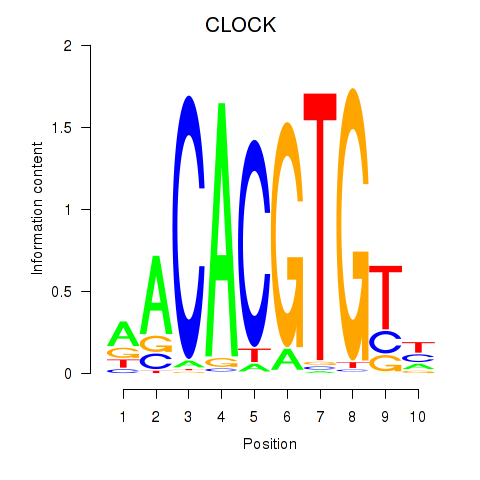
| CLOCK | 0.039 | 0.138 |
|
|
| CPEB1 | -0.060 | -0.231 |
|
|
| CREB1 | -0.018 | -0.062 |
|
|
| CREB3L1_CREB3 | 0.304 | 0.825 |
|
|
| CREB3L2 | 0.046 | 0.131 |
|
|
| CREB5_CREM_JUNB | -0.070 | -0.253 |
|
|
| CTCF_CTCFL | -0.128 | -0.517 |
|
|
| CUACAGU | 0.663 | 0.320 |
|

|
| CUGGCUG | 0.044 | 0.043 |
|

|
| CUUUGGU | -0.129 | -0.452 |
|

|
| CUX1 | -0.103 | -0.453 |
|
|
| CUX2 | -0.036 | -0.145 |
|
|
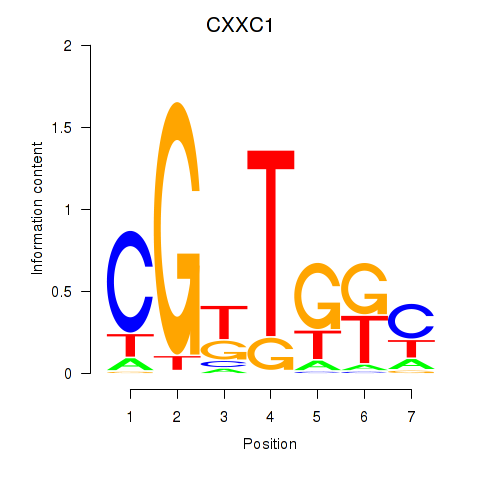
| CXXC1 | 0.113 | 0.687 |
|
|
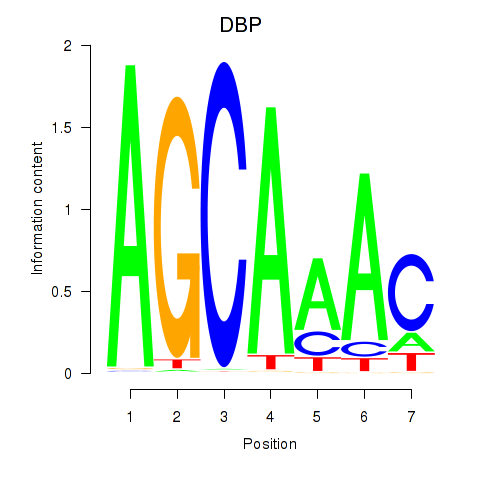
| DBP | 0.013 | 0.039 |
|
|
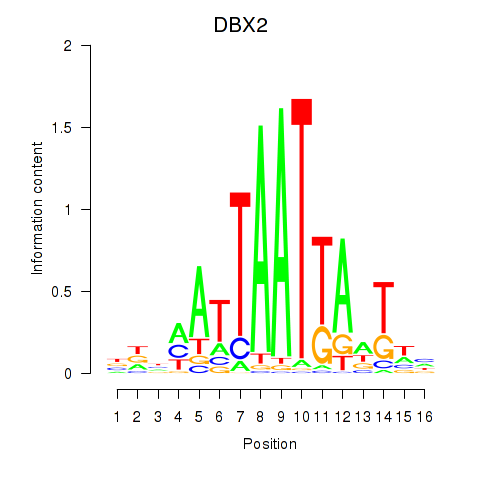
| DBX2_HLX | 0.001 | 0.003 |
|
|
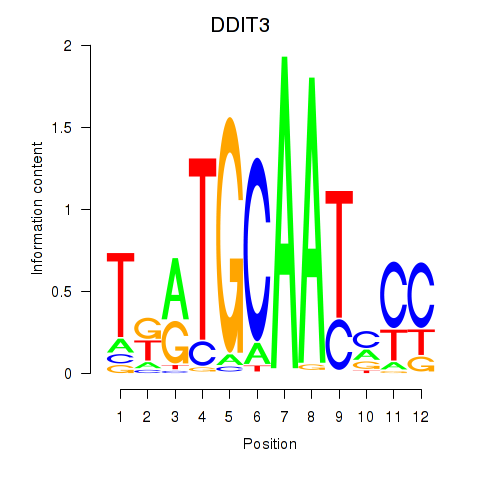
| DDIT3 | 0.141 | 0.222 |
|
|
| DLX1_HOXA3_BARX2 | 0.033 | 0.086 |
|
|
| DLX3_EVX1_MEOX1 | 0.217 | 0.360 |
|
|
| DLX4_HOXD8 | -0.122 | -0.626 |
|
|
| DLX5 | -0.056 | -0.074 |
|
|
| DMBX1 | -0.077 | -0.096 |
|
|
| DMC1 | 0.360 | 0.495 |
|

|
| DPRX | 0.175 | 0.540 |
|
|
| DRGX_PROP1 | -0.506 | -0.759 |
|
|
| DUXA | -0.257 | -0.871 |
|
|
| E2F2_E2F5 | -0.072 | -0.248 |
|
|
| E2F3 | 0.075 | 0.652 |
|
|
| E2F4 | -0.092 | -0.312 |
|
|
| E2F6 | -0.070 | -0.619 |
|
|
| E2F7_E2F1 | 0.050 | 0.356 |
|
|
| E2F8 | 0.062 | 0.209 |
|
|
| E4F1 | -0.210 | -0.337 |
|
|
| EBF1 | -0.075 | -0.652 |
|
|
| EBF3 | 0.089 | 0.348 |
|
|
| EGR1_EGR4 | -0.075 | -0.310 |
|
|
| EGR3_EGR2 | 0.089 | 0.356 |
|
|
| ELF2_GABPA_ELF5 | 0.057 | 0.628 |
|
|
| ELF3_EHF | -0.046 | -0.262 |
|
|
| ELK4_ETV5_ELK1_ELK3_ELF4 | 0.142 | 1.278 |
|
|
| EMX1 | -0.090 | -0.218 |
|
|
| EMX2 | -0.050 | -0.151 |
|

|
| EN1_ESX1_GBX1 | -0.247 | -0.568 |
|
|
| EN2_GBX2_LBX2 | 0.022 | 0.034 |
|
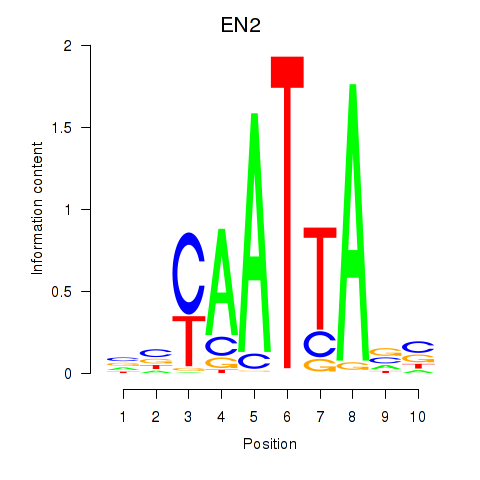
|
| EOMES | 0.139 | 0.561 |
|
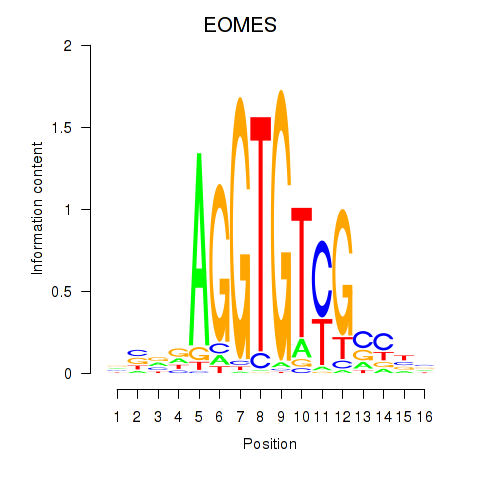
|
| EP300 | 0.004 | 0.014 |
|
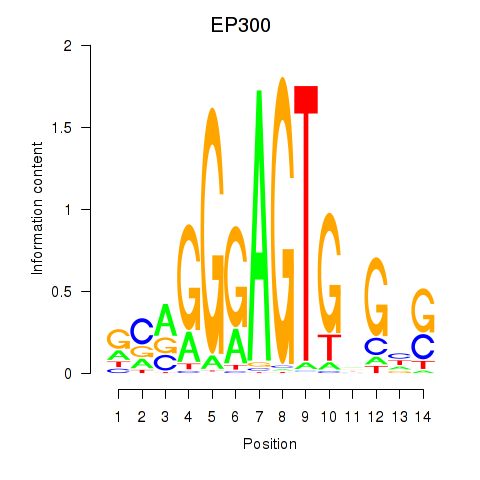
|
| EPAS1_BCL3 | -0.271 | -1.796 |
|
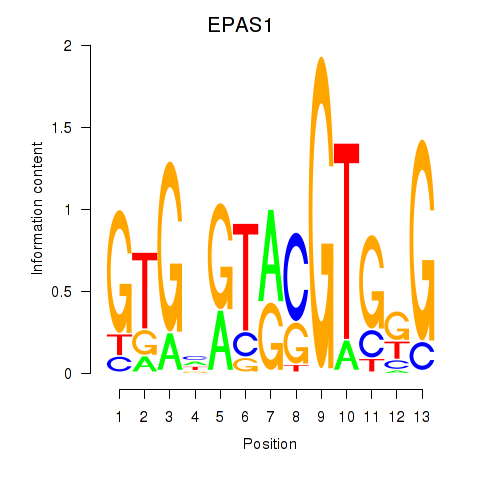
|
| ERG | -0.019 | -0.145 |
|
|
| ESR1 | -0.023 | -0.107 |
|

|
| ESRRA_ESR2 | -0.043 | -0.602 |
|
|
| ESRRB_ESRRG | 0.158 | 0.594 |
|
|
| ETS1 | 0.100 | 0.537 |
|

|
| ETV1_ERF_FEV_ELF1 | -0.015 | -0.094 |
|
|
| ETV2 | 0.104 | 0.503 |
|

|
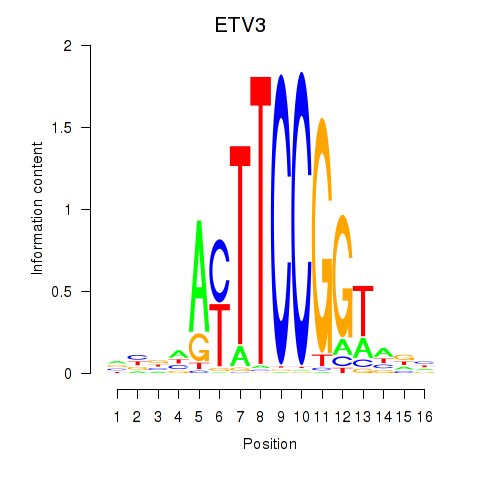
| ETV3 | -0.059 | -0.199 |
|
|
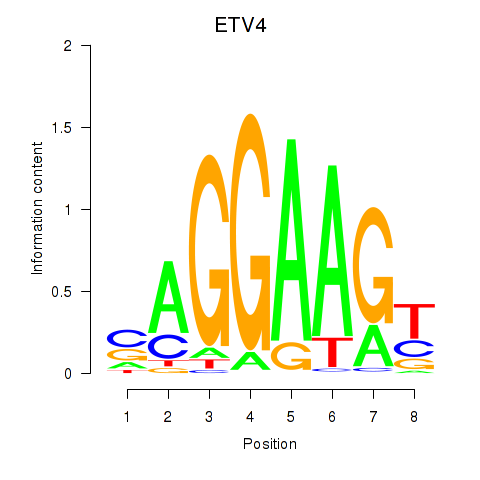
| ETV4_ETS2 | -0.108 | -0.503 |
|
|
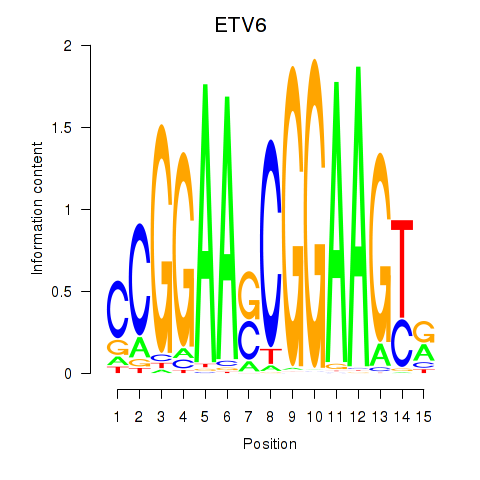
| ETV6 | 0.219 | 0.950 |
|
|
| ETV7 | 0.095 | 0.428 |
|

|
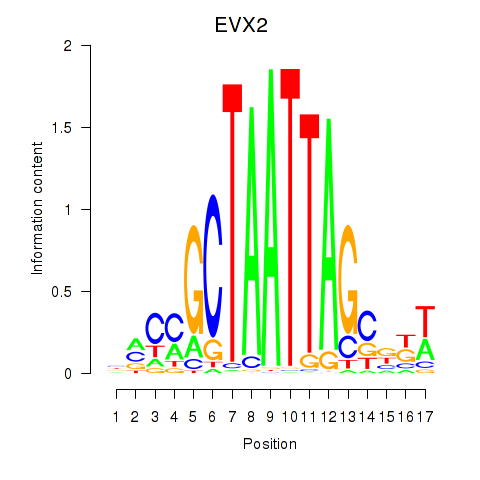
| EVX2 | -0.191 | -0.138 |
|
|
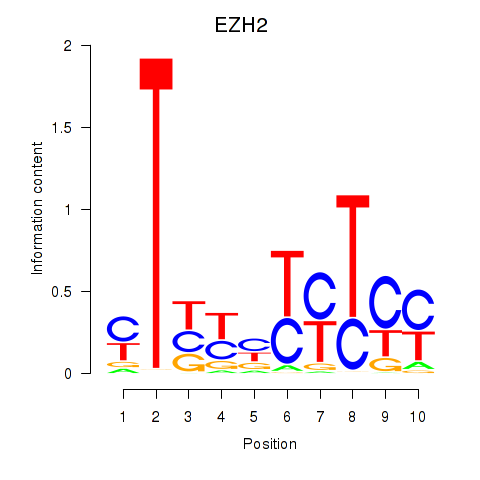
| EZH2 | -0.085 | -0.506 |
|
|
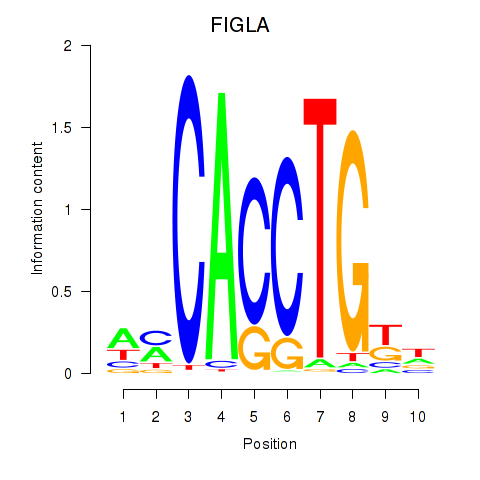
| FIGLA | -0.222 | -0.596 |
|
|
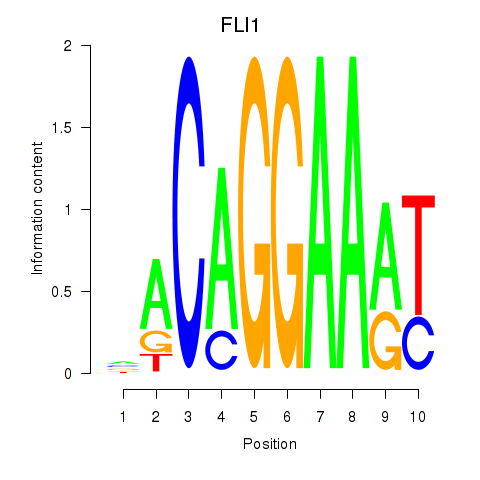
| FLI1 | -0.044 | -0.279 |
|
|
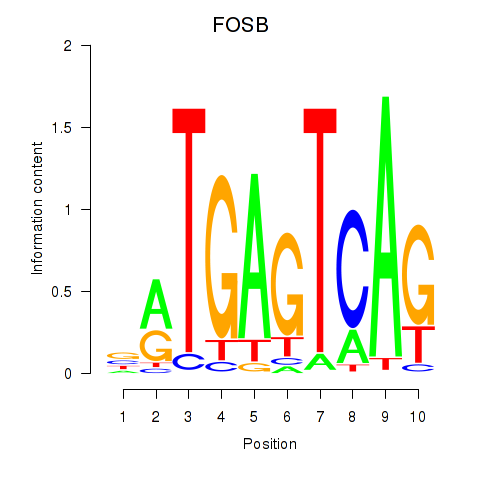
| FOSB | -0.334 | -0.808 |
|
|
| FOSL1 | 0.200 | 0.551 |
|
|
| FOSL2_SMARCC1 | -0.164 | -0.501 |
|
|
| FOXA1 | -0.320 | -0.736 |
|

|
| FOXA2_FOXJ3 | -0.053 | -0.168 |
|
|
| FOXA3_FOXC2 | 0.019 | 0.042 |
|
|
| FOXC1 | 0.233 | 0.605 |
|
|
| FOXD1_FOXO1_FOXO6_FOXG1_FOXP1 | 0.059 | 0.223 |
|
|
| FOXD3_FOXI1_FOXF1 | -0.009 | -0.037 |
|
|
| FOXF2_FOXJ1 | 0.174 | 0.399 |
|
|
| FOXJ2 | 0.067 | 0.140 |
|
|
| FOXK1_FOXP2_FOXB1_FOXP3 | 0.077 | 0.304 |
|
|
| FOXL1 | -0.126 | -0.741 |
|
|
| FOXM1_TBL1XR1 | -0.017 | -0.264 |
|
|
| FOXN1 | 0.019 | 0.057 |
|
|
| FOXO3_FOXD2 | -0.215 | -0.581 |
|
|
| FOXO4 | -0.173 | -0.484 |
|
|
| FOXQ1 | 0.090 | 0.326 |
|
|
| FUBP1 | 0.051 | 0.103 |
|
|
| GAGAACU | -1.199 | -0.321 |
|

|
| GAGAUGA | -0.121 | -0.129 |
|

|
| GAGGUAG | -0.130 | -0.464 |
|

|
| GATA1_GATA4 | -0.122 | -0.489 |
|
|
| GATA2 | 0.015 | 0.072 |
|
|
| GATA3 | 0.012 | 0.162 |
|
|
| GATA5 | -0.318 | -0.685 |
|
|
| GATA6 | -0.266 | -0.404 |
|

|
| GAUAUGU | -0.989 | -0.585 |
|

|
| GAUCAGA | 2.735 | 0.623 |
|

|
| GAUUGUC | 0.010 | 0.020 |
|

|
| GCAGCAU | -0.224 | -0.450 |
|

|
| GCCUGUC | -0.011 | -0.006 |
|

|
| GCM1 | -0.290 | -0.758 |
|
|
| GCM2 | -0.172 | -0.431 |
|

|
| GCUACAU | -0.400 | -0.577 |
|

|
| GCUGGUG | -0.143 | -0.329 |
|

|
| GFI1 | -0.243 | -0.980 |
|
|
| GFI1B | -0.136 | -0.444 |
|
|
| GGAAGAC | 0.212 | 0.338 |
|

|
| GGAAUGU | 0.466 | 1.247 |
|

|
| GGACGGA | -1.145 | -0.374 |
|

|
| GGAGUGU | 0.684 | 0.387 |
|

|
| GGCAAGA | 0.259 | 0.289 |
|

|
| GGCAGUG | 0.148 | 0.360 |
|

|
| GGCUCAG | -0.296 | -0.542 |
|

|
| GLI2 | -0.027 | -0.127 |
|

|
| GLI3 | 0.035 | 0.208 |
|
|
| GLIS1 | -0.211 | -0.131 |
|
|
| GLIS2 | 0.194 | 0.642 |
|
|
| GLIS3 | -0.233 | -0.326 |
|
|
| GMEB1 | -0.036 | -0.244 |
|
|
| GMEB2 | 0.287 | 1.464 |
|
|
| GRHL1 | -0.226 | -0.302 |
|
|
| GSC_GSC2 | -0.323 | -0.614 |
|
|
| GTF2I | -0.000 | -0.002 |
|
|
| GUAAACA | -0.069 | -0.253 |
|

|
| GUAACAG | 0.253 | 0.315 |
|

|
| GUACCGU | -0.097 | -0.016 |
|

|
| GUAGUGU | -0.068 | -0.115 |
|

|
| GUCAGUU | -0.195 | -0.210 |
|

|
| GUGCAAA | 0.044 | 0.127 |
|

|
| GZF1 | -0.039 | -0.048 |
|
|
| HAND1 | -0.380 | -0.659 |
|

|
| HBP1 | -0.173 | -0.500 |
|
|
| HDX | -0.974 | -0.824 |
|
|
| HES1 | 0.144 | 0.743 |
|

|
| HES7_HES5 | -0.027 | -0.066 |
|
|
| HESX1 | -0.281 | -0.585 |
|
|
| HIC1 | 0.005 | 0.035 |
|
|
| HIC2 | -0.141 | -1.119 |
|
|
| HIF1A | 0.000 | 0.000 |
|
|
| HINFP | -0.015 | -0.042 |
|
|
| HINFP1 | 0.119 | 0.125 |
|
|
| HIVEP1 | -0.081 | -0.292 |
|
|
| HLF_TEF | 0.189 | 0.423 |
|
|
| HMBOX1 | 0.183 | 0.450 |
|
|
| HMGA1 | 0.066 | 0.281 |
|
|
| HMGA2 | 0.259 | 0.373 |
|
|
| HMX1 | -0.033 | -0.105 |
|

|
| HMX2 | 0.017 | 0.034 |
|
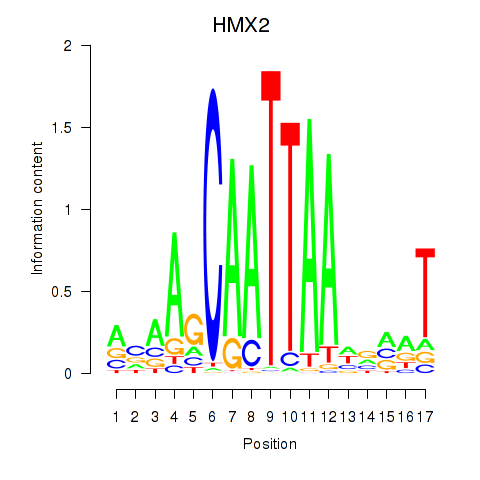
|
| HMX3 | -0.191 | -0.827 |
|
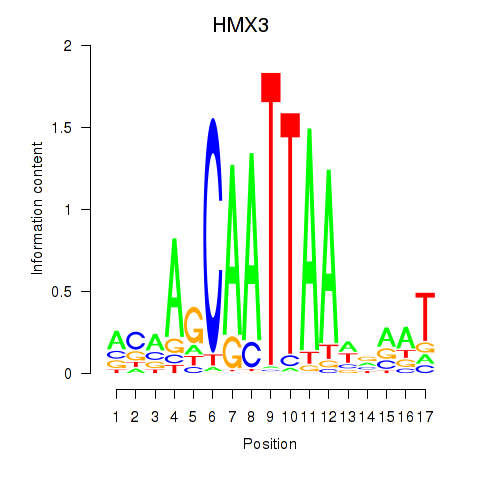
|
| HNF1A_HNF1B | -0.292 | -0.737 |
|
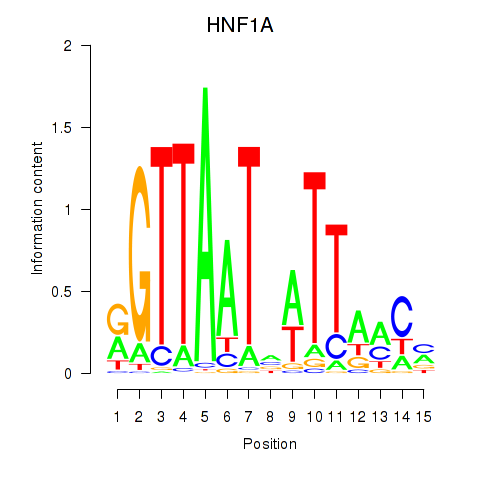
|
| HNF4A | -0.131 | -0.300 |
|
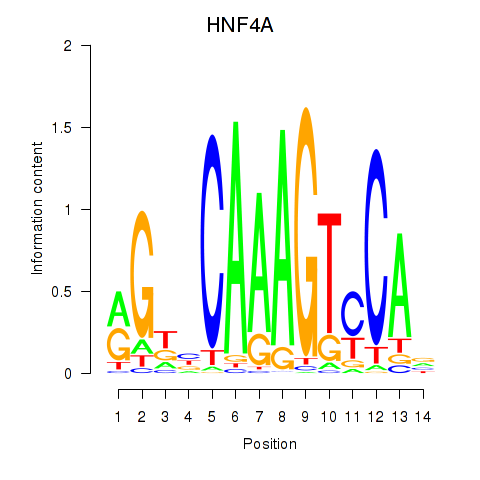
|
| HNF4G | -0.074 | -0.311 |
|
|
| HOMEZ | 0.095 | 0.391 |
|
|
| HOXA1 | -0.056 | -0.169 |
|
|
| HOXA10_HOXB9 | -0.452 | -0.881 |
|
|
| HOXA13 | -0.196 | -0.505 |
|
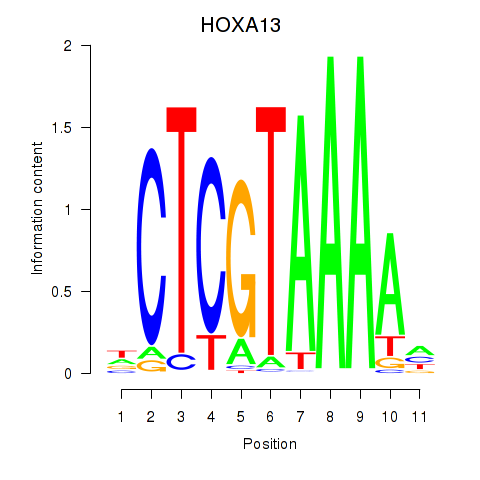
|
| HOXA2_HOXB1 | -0.307 | -0.561 |
|
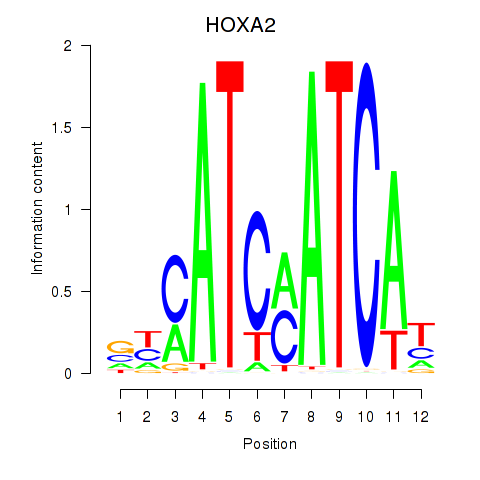
|
| HOXA4 | -0.189 | -0.350 |
|
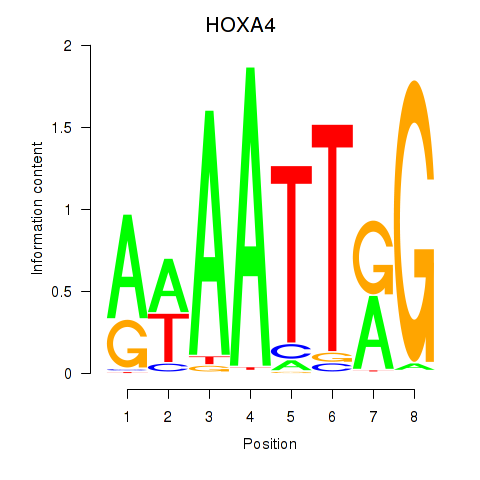
|
| HOXA5 | -0.248 | -0.425 |
|
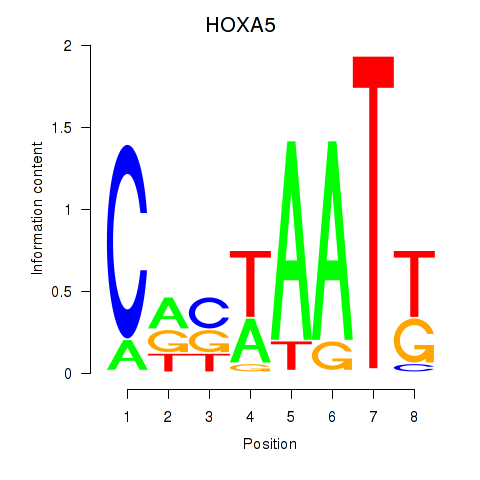
|
| HOXA6 | 0.125 | 0.216 |
|
|
| HOXA9 | -0.018 | -0.056 |
|
|
| HOXB13 | -0.077 | -0.090 |
|
|
| HOXB2_UNCX_HOXD3 | 0.113 | 0.394 |
|
|
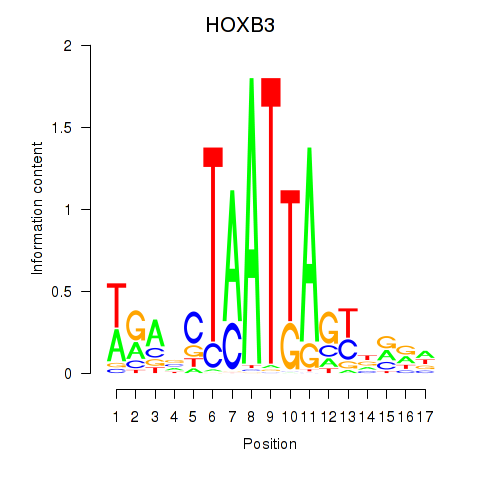
| HOXB3 | -0.171 | -0.399 |
|
|
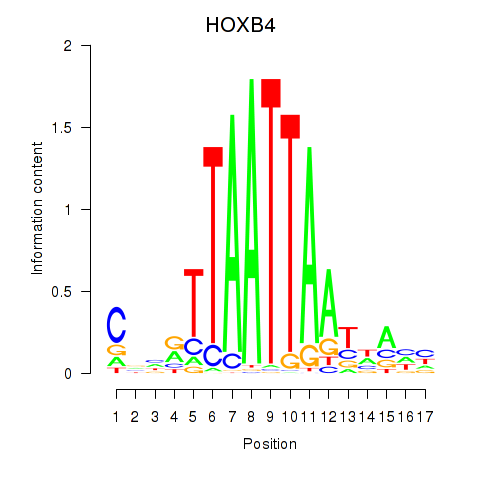
| HOXB4_LHX9 | -0.152 | -0.329 |
|
|
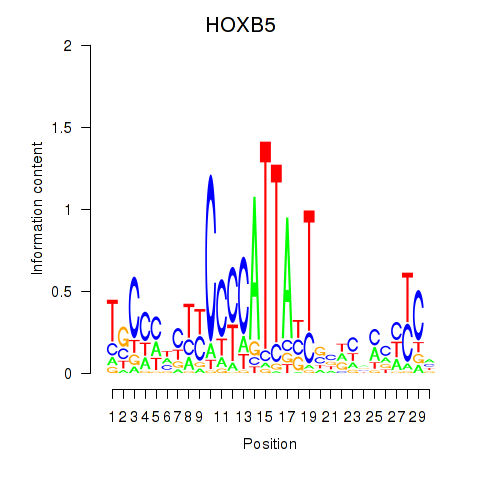
| HOXB5 | -0.230 | -0.343 |
|
|
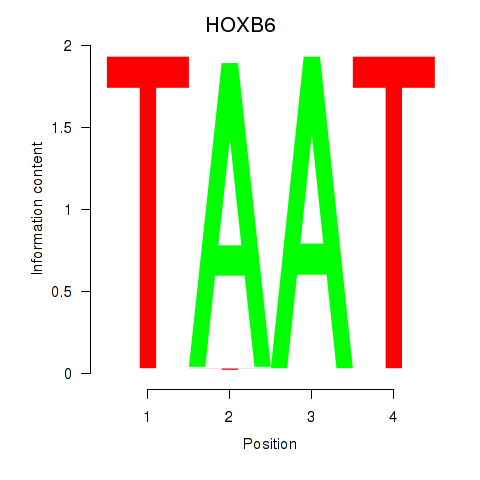
| HOXB6_PRRX2 | -0.232 | -0.985 |
|
|
| HOXB7 | -0.251 | -0.677 |
|
|
| HOXB8 | 0.025 | 0.108 |
|
|
| HOXC10_HOXD13 | 0.121 | 0.402 |
|
|
| HOXC11 | -0.143 | -0.262 |
|
|
| HOXC12_HOXD12 | 0.105 | 0.311 |
|

|
| HOXC13 | -0.237 | -0.546 |
|
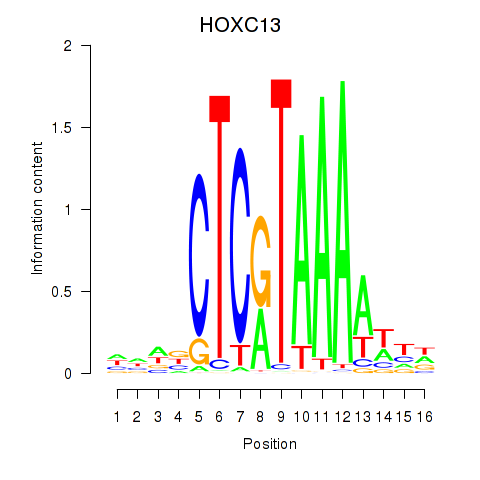
|
| HOXC6_HOXA7 | -0.133 | -0.195 |
|
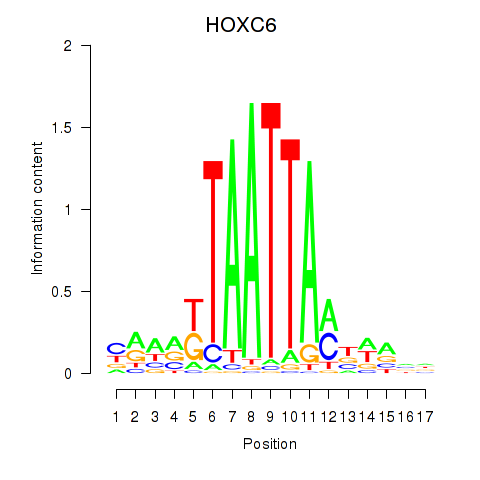
|
| HOXC8 | 0.143 | 0.899 |
|
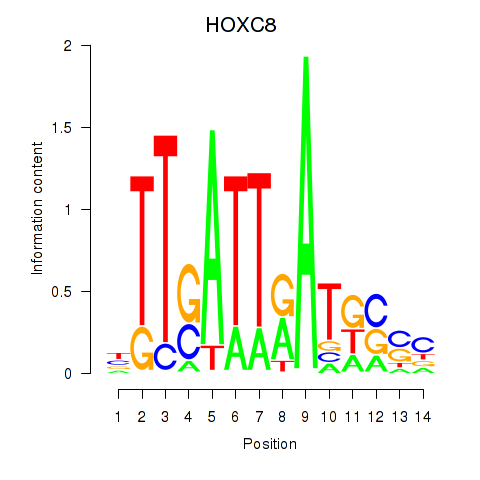
|
| HOXC9 | -0.083 | -0.215 |
|
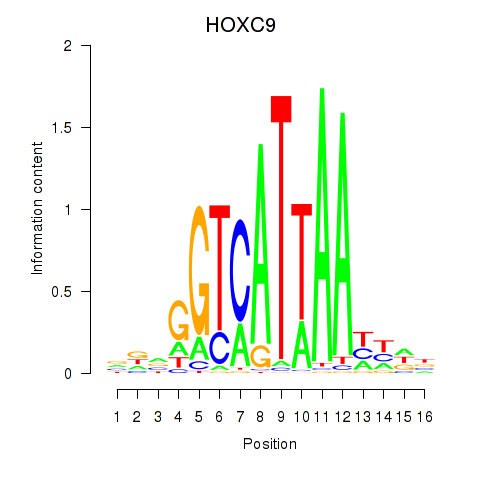
|
| HOXD1 | 0.540 | 0.545 |
|

|
| HOXD10 | -0.096 | -0.289 |
|
|
| HOXD11_HOXA11 | 0.070 | 0.220 |
|
|
| HOXD4 | -0.061 | -0.117 |
|
|
| HOXD9 | -0.072 | -0.171 |
|
|
| HSF1 | -0.005 | -0.021 |
|

|
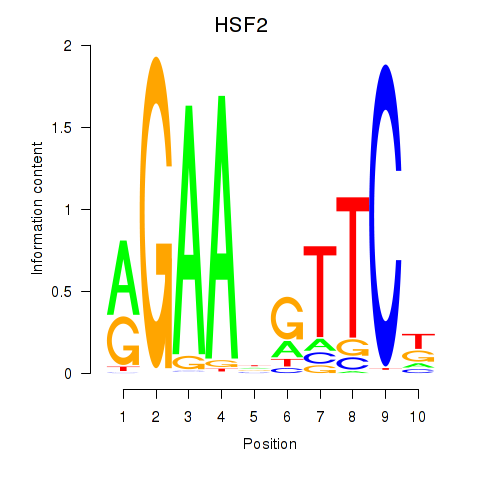
| HSF2 | 0.038 | 0.077 |
|
|
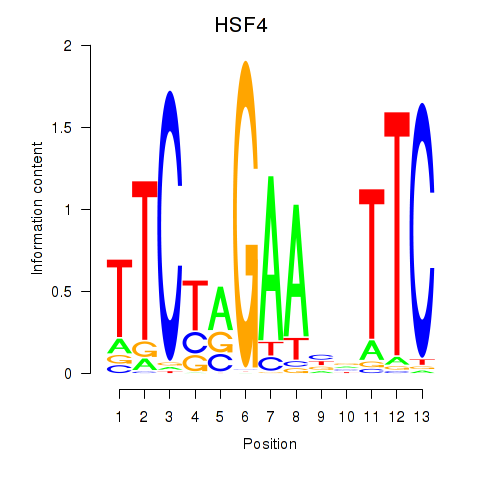
| HSF4 | 0.499 | 1.345 |
|
|
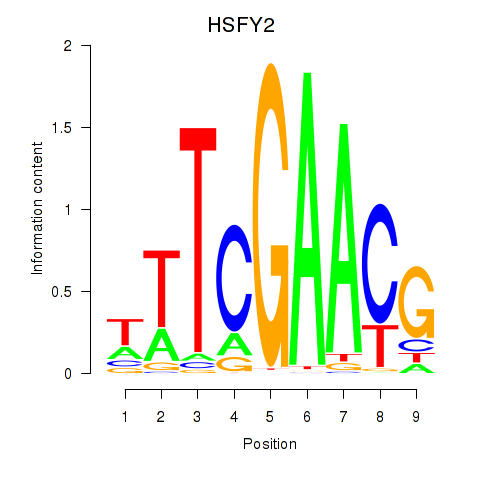
| HSFY2 | -0.041 | -0.169 |
|
|
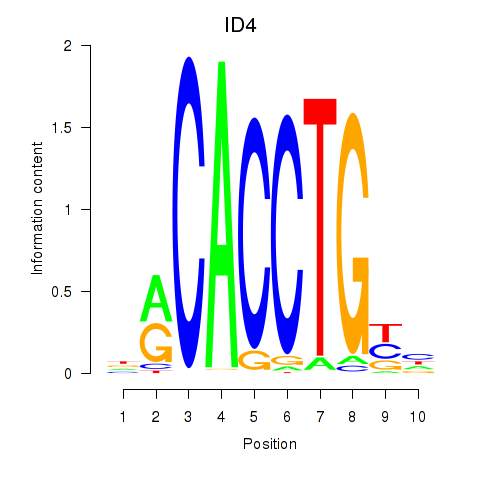
| ID4_TCF4_SNAI2 | -0.075 | -0.745 |
|
|
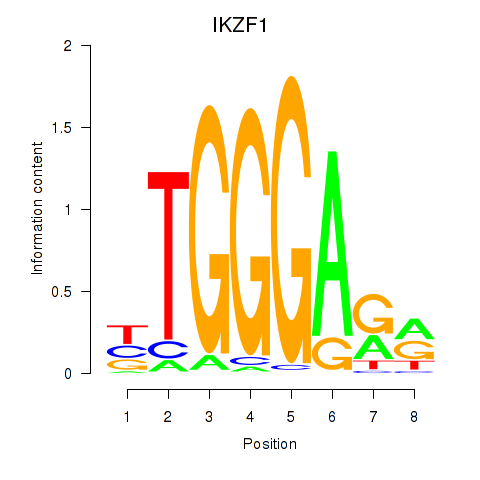
| IKZF1 | -0.008 | -0.153 |
|
|
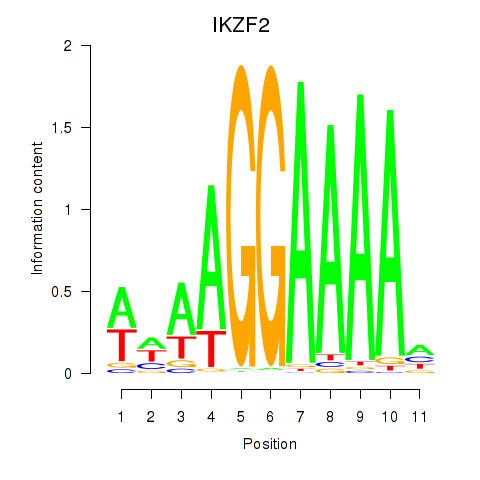
| IKZF2 | 0.284 | 0.936 |
|
|
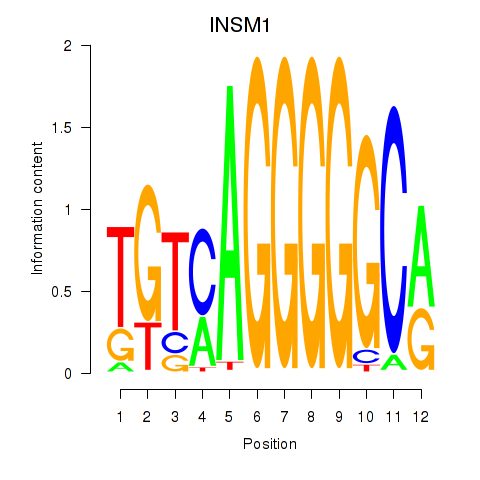
| INSM1 | -0.212 | -0.519 |
|
|
| IRF2_STAT2_IRF8_IRF1 | -0.055 | -0.241 |
|

|
| IRF3 | 0.127 | 0.295 |
|

|
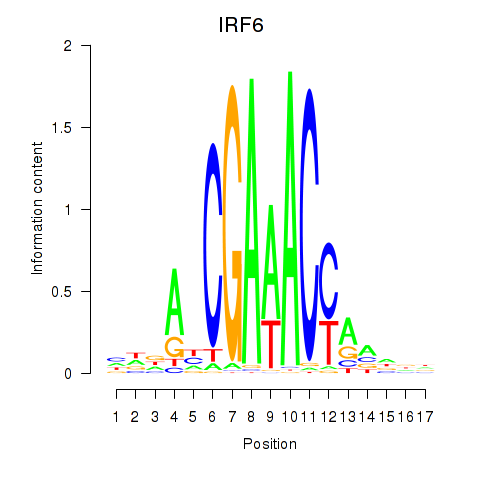
| IRF6_IRF4_IRF5 | 0.077 | 0.184 |
|
|
| IRF7 | -0.053 | -0.240 |
|
|
| IRF9 | 0.252 | 0.412 |
|

|
| IRX2 | 0.074 | 0.143 |
|
|
| IRX3 | -0.194 | -0.763 |
|
|
| IRX5 | 0.289 | 0.537 |
|
|
| IRX6_IRX4 | 0.524 | 0.501 |
|
|
| ISL1 | -0.315 | -0.735 |
|
|
| ISL2 | -0.135 | -0.219 |
|
|
| ISX | -0.307 | -0.612 |
|
|
| JDP2 | -0.073 | -0.161 |
|
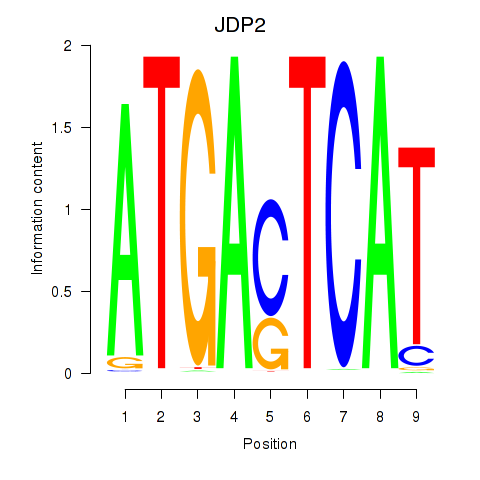
|
| JUN | -0.002 | -0.006 |
|

|
| JUND | -0.092 | -0.181 |
|
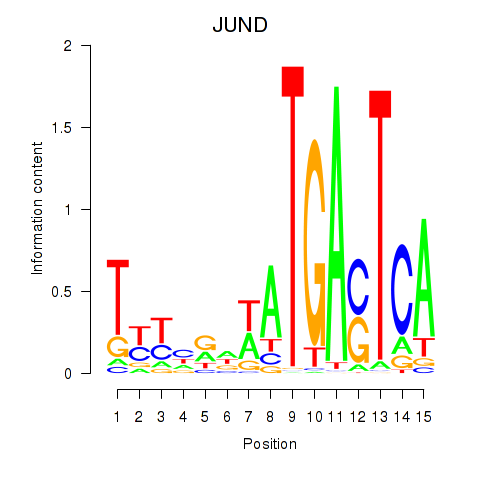
|
| KLF1 | 0.313 | 1.213 |
|
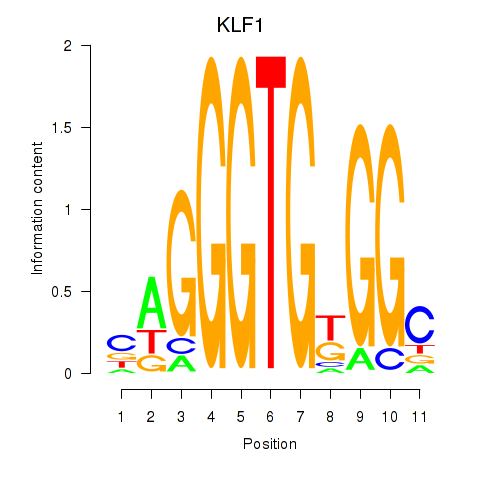
|
| KLF12 | 0.109 | 0.400 |
|
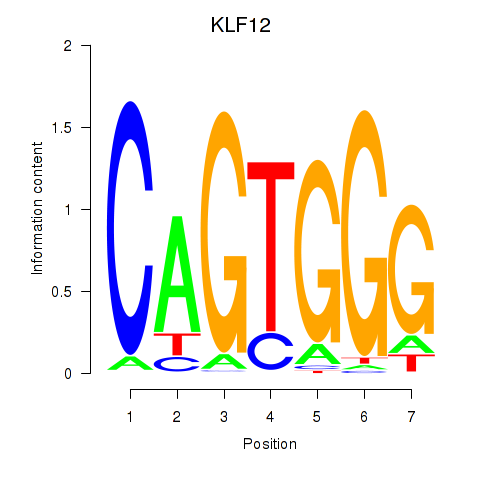
|
| KLF13 | -0.121 | -0.337 |
|
|
| KLF14_SP8 | -0.057 | -0.515 |
|
|
| KLF15 | 0.076 | 0.260 |
|
|
| KLF16_SP2 | 0.022 | 0.311 |
|
|
| KLF3 | -0.163 | -0.683 |
|

|
| KLF6 | -0.087 | -0.339 |
|
|
| KLF7 | -0.126 | -0.141 |
|
|
| KLF8 | -0.033 | -0.371 |
|
|
| LEF1 | -0.241 | -0.931 |
|
|
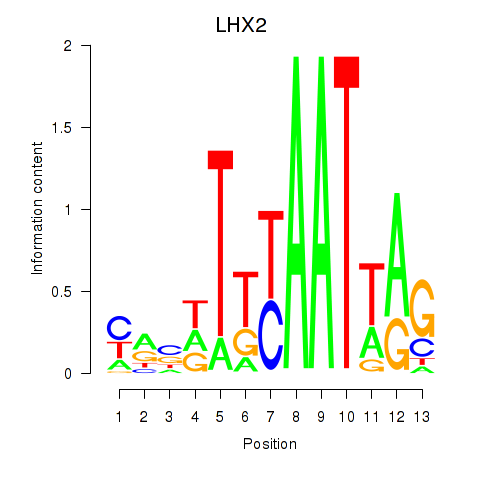
| LHX2 | 0.232 | 0.414 |
|
|
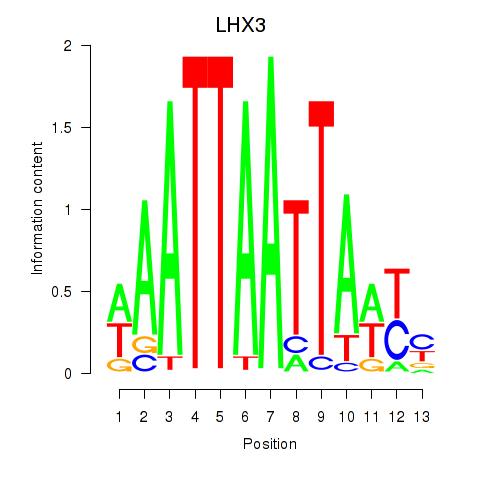
| LHX3 | 0.234 | 0.380 |
|
|
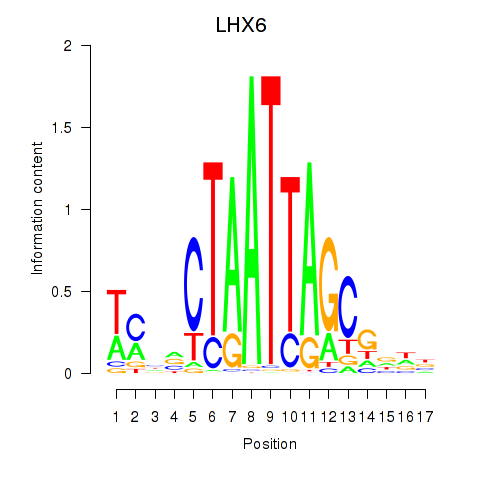
| LHX6 | -0.100 | -0.235 |
|
|
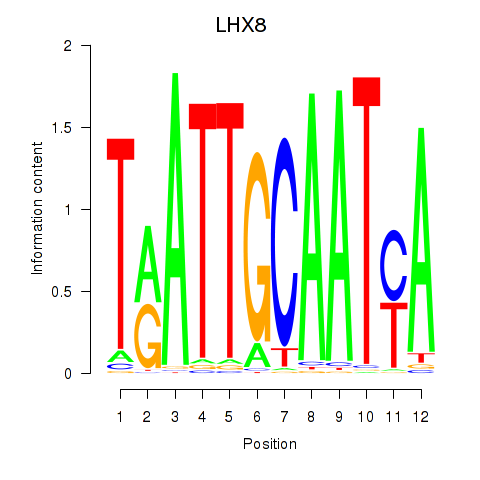
| LHX8 | 0.125 | 0.320 |
|
|
| LMX1B_MNX1_RAX2 | -0.297 | -0.493 |
|
|
| MAFA | 0.363 | 0.662 |
|
|
| MAFB | 0.056 | 0.478 |
|
|
| MAFF_MAFG | 0.031 | 0.105 |
|
|
| MAFK | 0.038 | 0.240 |
|
|
| MAF_NRL | -0.136 | -0.545 |
|
|
| MAX_TFEB | 0.132 | 0.443 |
|
|
| MAZ_ZNF281_GTF2F1 | -0.016 | -0.227 |
|
|
| MBD2 | -0.319 | -1.183 |
|

|
| MECOM | -0.326 | -0.796 |
|
|
| MECP2 | 0.007 | 0.141 |
|
|
| MEF2B | -0.286 | -0.483 |
|
|
| MEF2C | 0.018 | 0.038 |
|
|
| MEF2D_MEF2A | 0.058 | 0.127 |
|
|
| MEIS1 | -0.081 | -0.404 |
|
|
| MEIS2 | -0.009 | -0.091 |
|
|
| MEIS3_TGIF2LX | -0.144 | -0.334 |
|
|
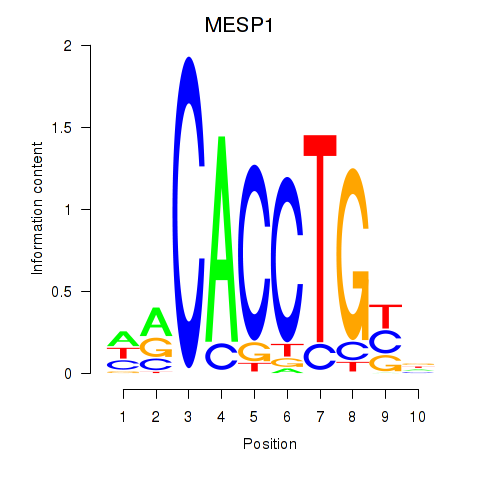
| MESP1 | 0.073 | 0.340 |
|
|
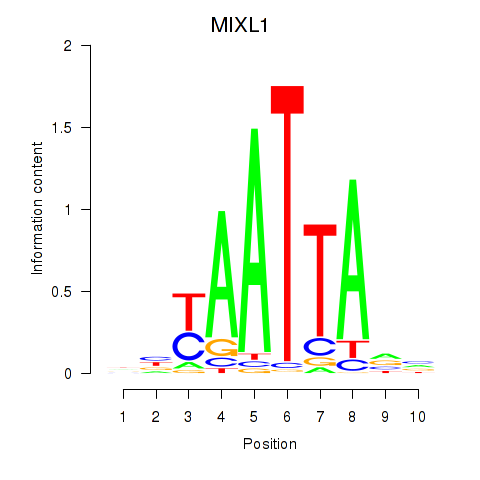
| MIXL1_GSX1_BSX_MEOX2_LHX4 | 0.051 | 0.162 |
|
|
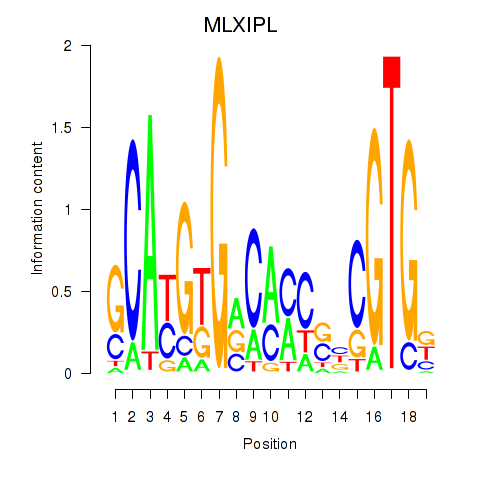
| MLXIPL | -0.015 | -0.134 |
|
|
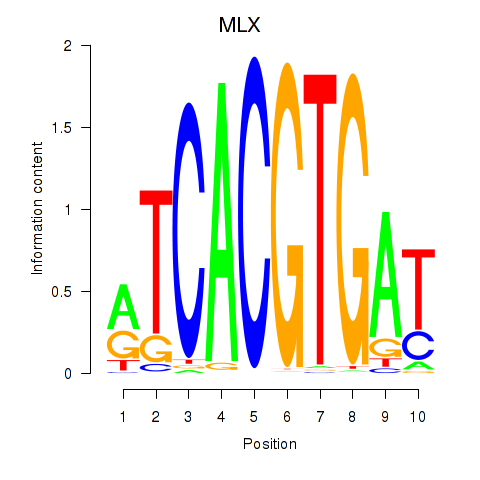
| MLX_USF2_USF1_PAX2 | -0.064 | -0.260 |
|
|
| MNT_HEY1_HEY2 | -0.108 | -0.598 |
|
|
| MSX1 | 0.116 | 0.234 |
|
|
| MSX2 | -0.107 | -0.162 |
|
|
| MXI1_MYC_MYCN | 0.103 | 0.767 |
|
|
| MYB | 0.121 | 0.699 |
|
|
| MYBL1 | -0.006 | -0.019 |
|
|
| MYBL2 | -0.061 | -0.267 |
|
|
| MYF6 | -0.015 | -0.090 |
|

|
| MYOD1 | -0.080 | -0.292 |
|
|
| MZF1 | 0.012 | 0.068 |
|
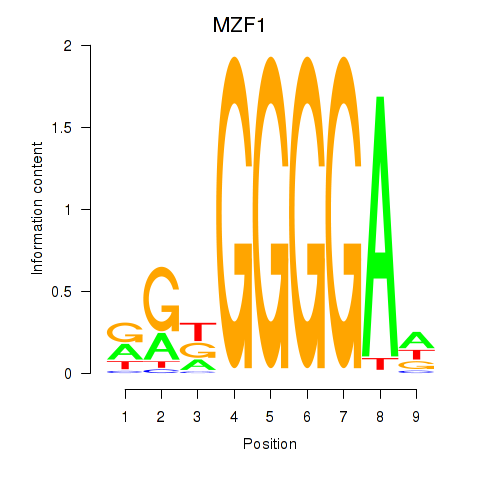
|
| NANOG | 0.267 | 0.563 |
|
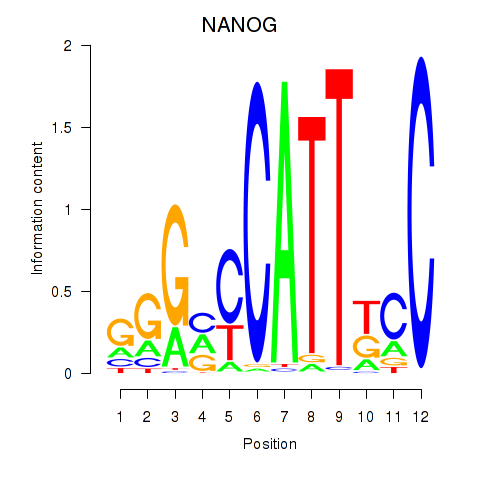
|
| NFAT5 | -0.019 | -0.039 |
|
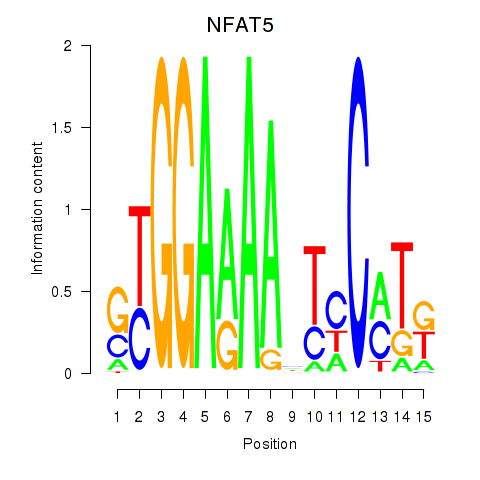
|
| NFATC1 | 0.086 | 0.284 |
|
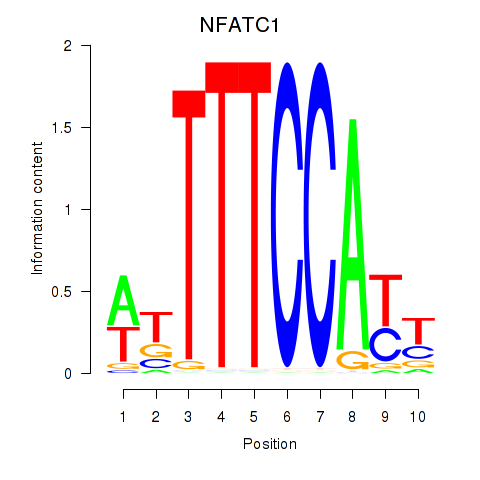
|
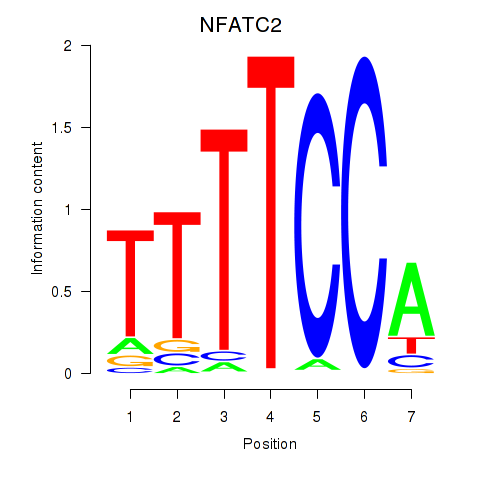
| NFATC2_NFATC3 | -0.025 | -0.107 |
|
|
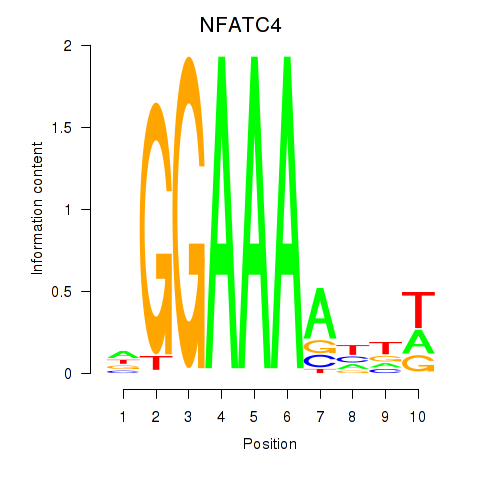
| NFATC4 | -0.295 | -0.973 |
|
|
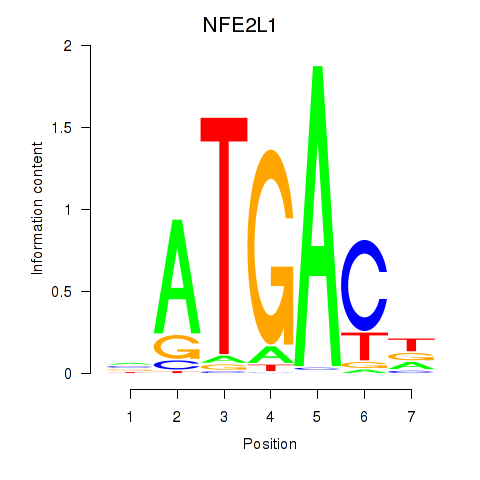
| NFE2L1 | -0.154 | -0.566 |
|
|
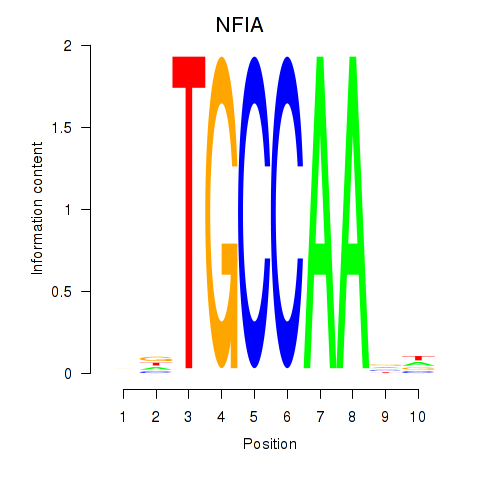
| NFIA | -0.071 | -0.269 |
|
|
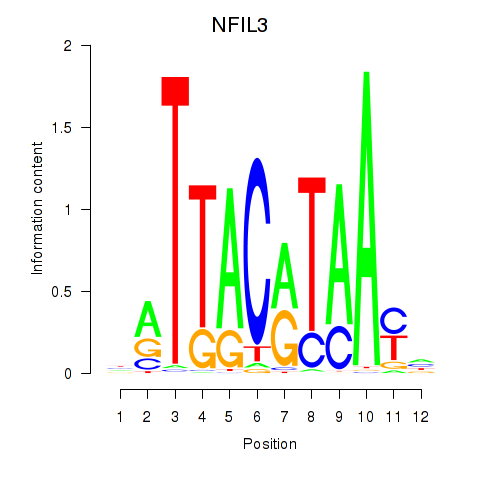
| NFIL3 | -0.013 | -0.072 |
|
|
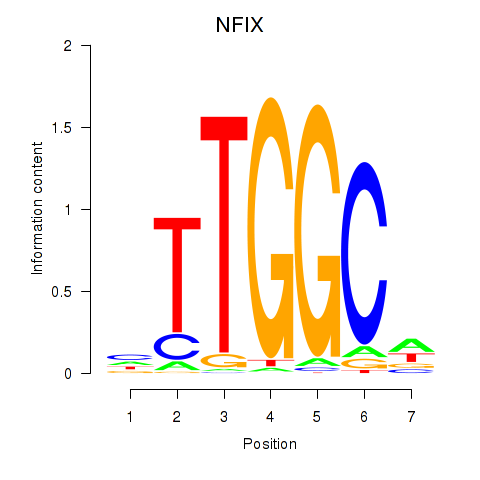
| NFIX_NFIB | -0.006 | -0.062 |
|
|
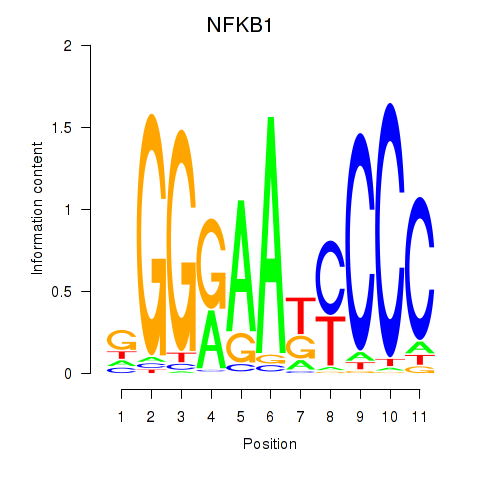
| NFKB1 | 0.055 | 0.141 |
|
|
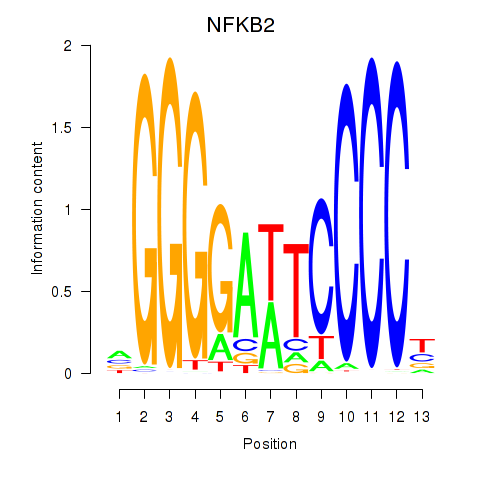
| NFKB2 | -0.061 | -0.237 |
|
|
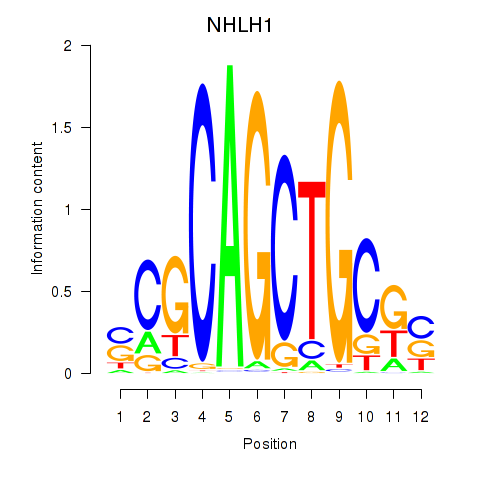
| NHLH1 | 0.008 | 0.035 |
|
|
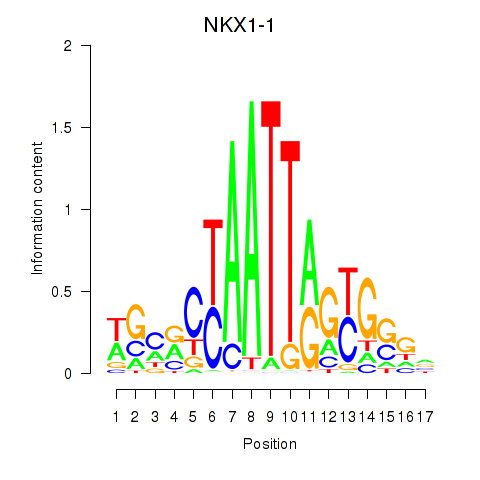
| NKX1-1 | -0.179 | -0.251 |
|
|
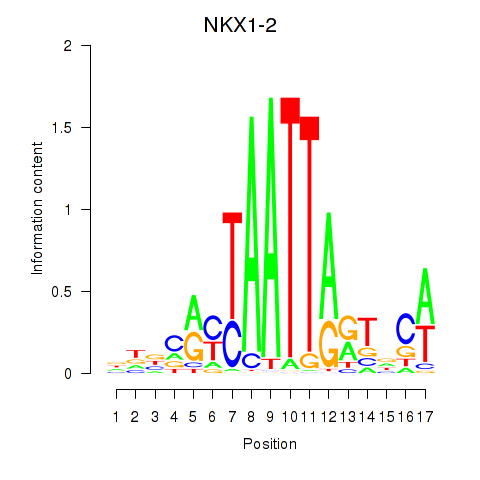
| NKX1-2_RAX | 0.419 | 0.599 |
|
|
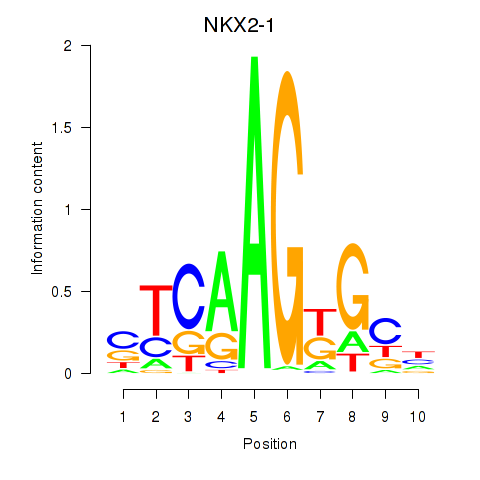
| NKX2-1 | -0.113 | -0.223 |
|
|
| NKX2-2 | 0.126 | 0.385 |
|
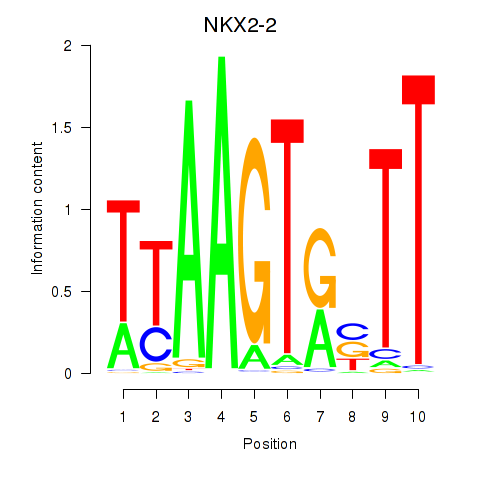
|
| NKX2-3 | 0.096 | 0.299 |
|
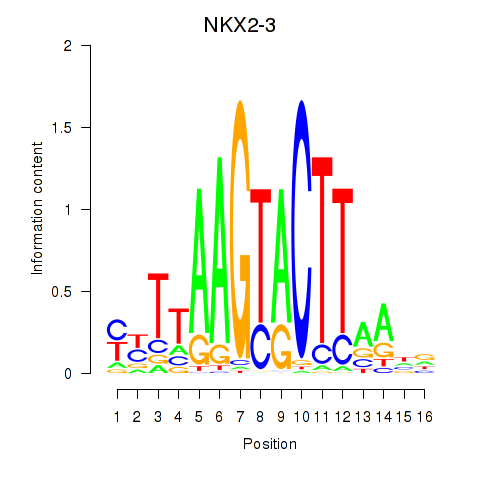
|
| NKX2-4 | -0.043 | -0.138 |
|
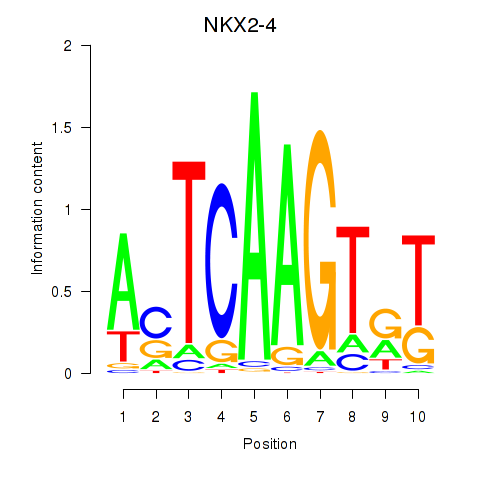
|
| NKX2-5 | -0.010 | -0.021 |
|
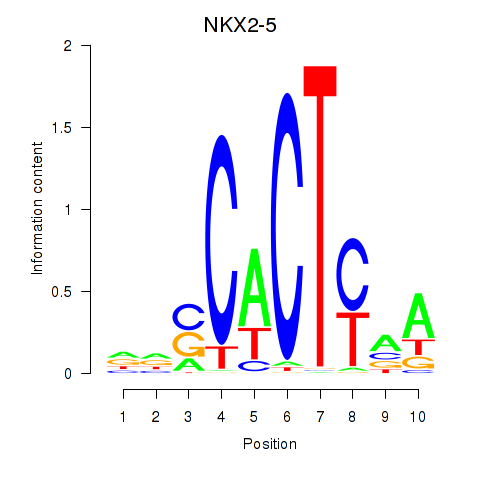
|
| NKX2-6 | -0.004 | -0.009 |
|
|
| NKX2-8 | -0.034 | -0.080 |
|
|
| NKX3-1 | 0.048 | 0.128 |
|
|
| NKX3-2 | -0.150 | -0.479 |
|
|
| NKX6-2 | 0.078 | 0.179 |
|
|
| NKX6-3 | 0.072 | 0.058 |
|
|
| NOTO_VSX2_DLX2_DLX6_NKX6-1 | 0.325 | 0.645 |
|
|
| NR0B1 | -0.018 | -0.053 |
|

|
| NR1D1 | -0.221 | -0.380 |
|
|
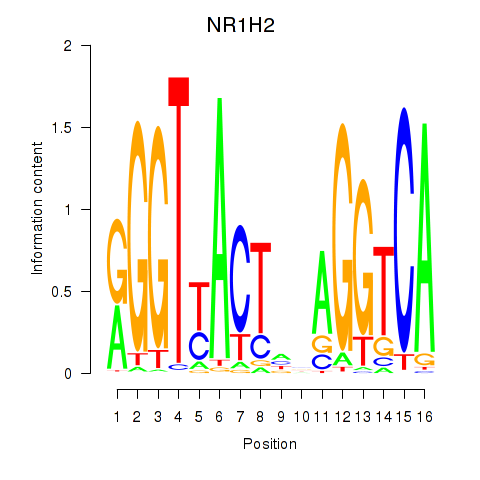
| NR1H2 | -0.015 | -0.037 |
|
|
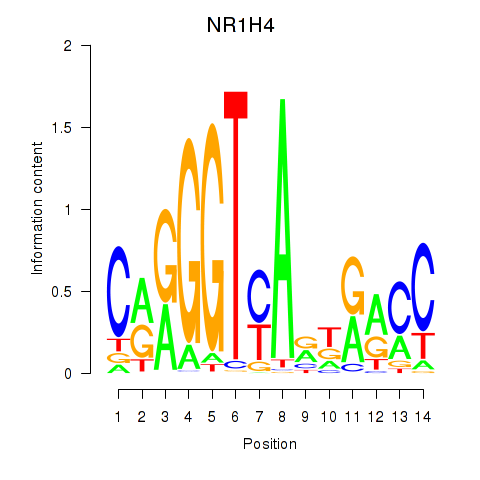
| NR1H4 | -0.232 | -0.554 |
|
|
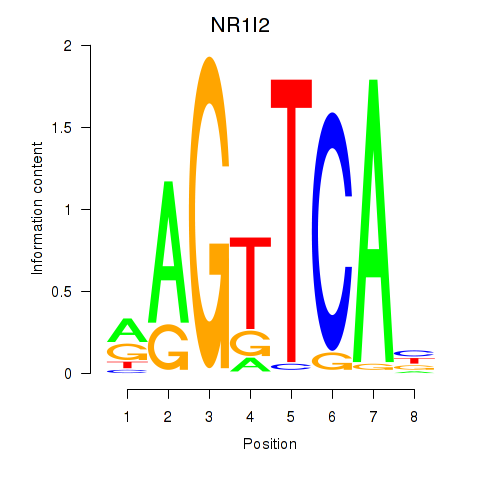
| NR1I2 | 0.094 | 0.300 |
|
|
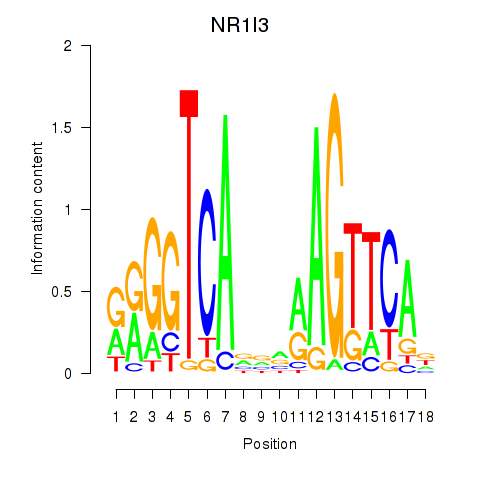
| NR1I3 | -0.058 | -0.090 |
|
|
| NR2C1 | 0.159 | 0.299 |
|
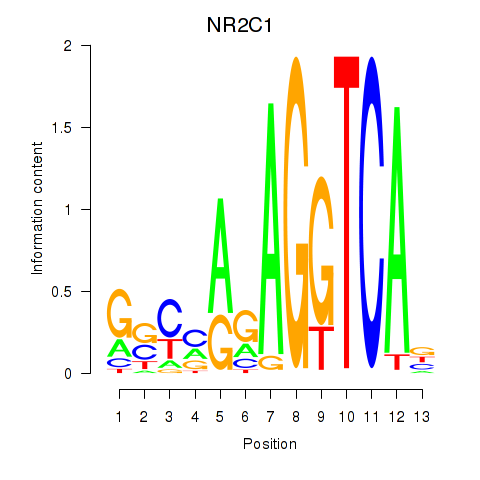
|
| NR2E1 | -0.192 | -0.579 |
|
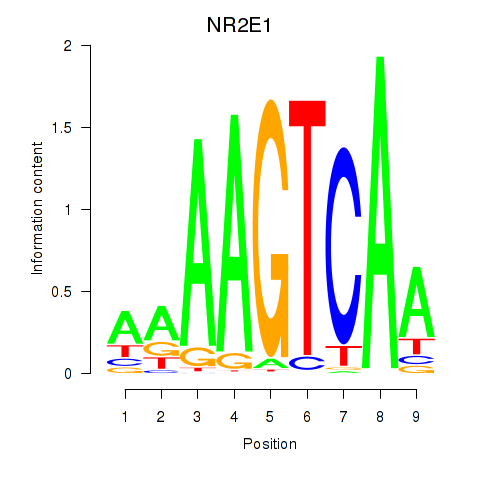
|
| NR2E3 | 0.026 | 0.104 |
|
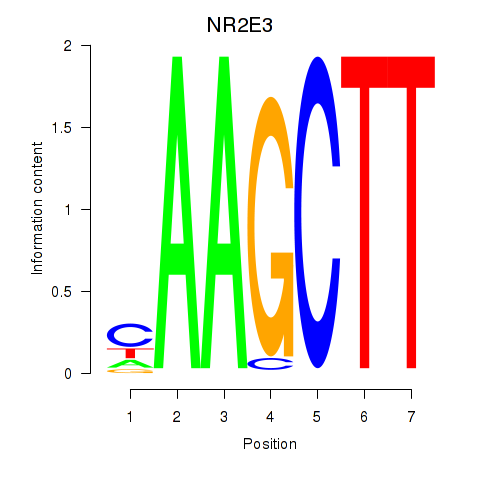
|
| NR2F1 | -0.151 | -0.403 |
|
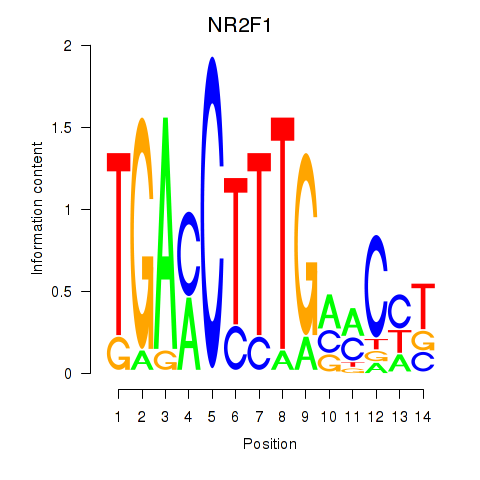
|
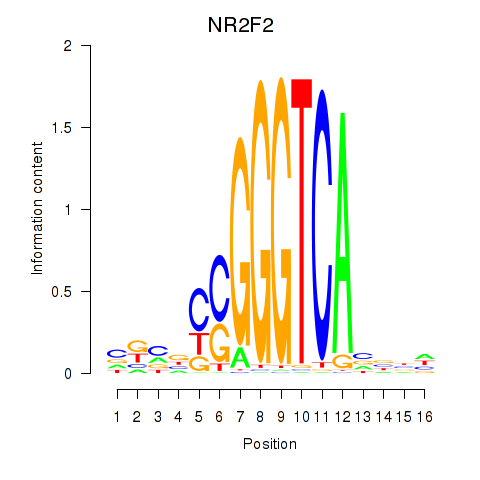
| NR2F2 | 0.143 | 0.616 |
|
|
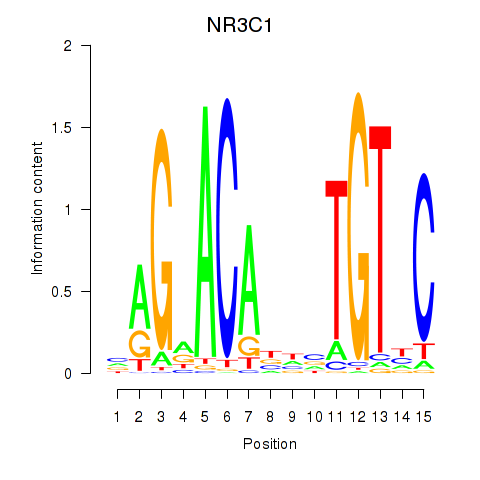
| NR3C1 | 0.020 | 0.116 |
|
|
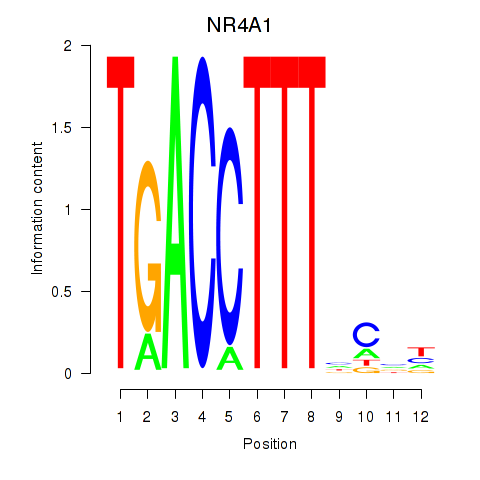
| NR4A1 | -0.222 | -0.429 |
|
|
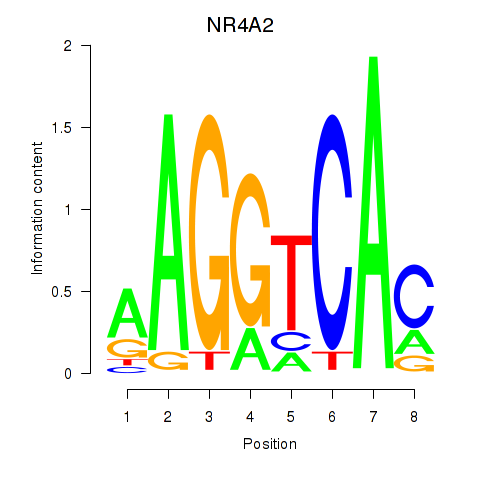
| NR4A2 | 0.017 | 0.037 |
|
|
| NR4A3 | 0.112 | 0.427 |
|
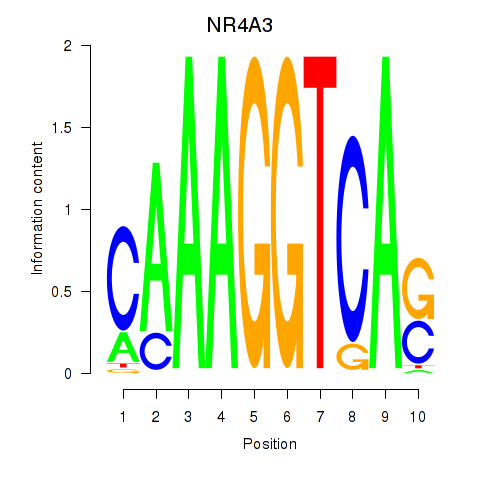
|
| NR5A2 | -0.035 | -0.158 |
|
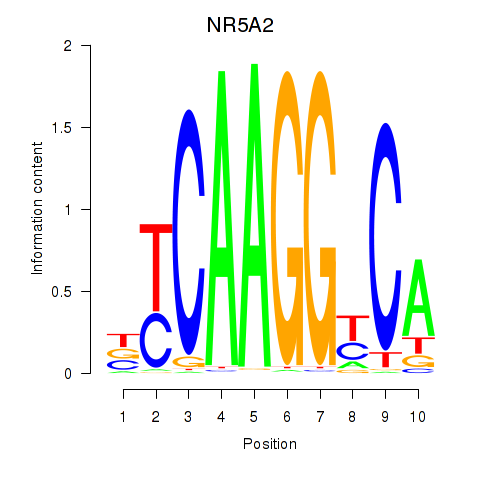
|
| NR6A1 | -0.140 | -0.453 |
|
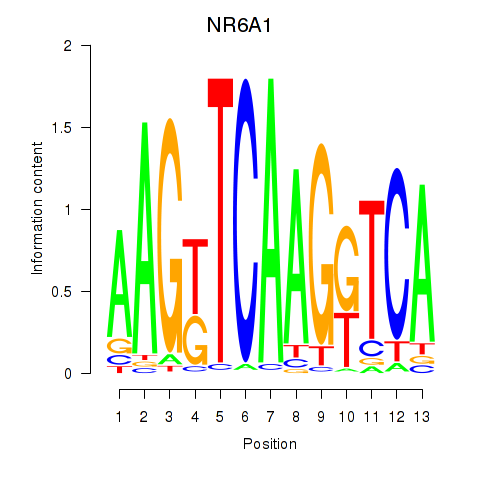
|
| NRF1 | 0.196 | 2.445 |
|
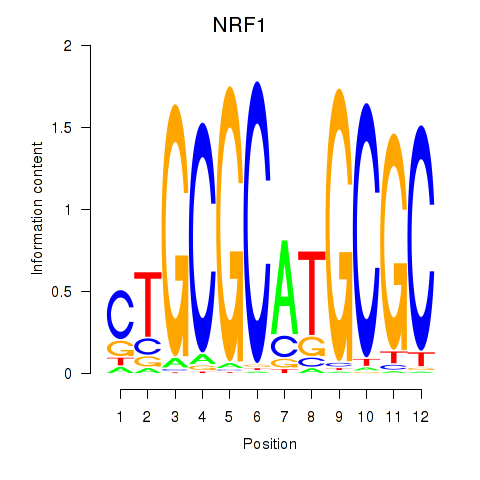
|
| OLIG1 | -0.113 | -0.240 |
|
|
| OLIG2_NEUROD1_ATOH1 | 0.076 | 0.285 |
|
|
| OLIG3_NEUROD2_NEUROG2 | -0.026 | -0.075 |
|
|
| ONECUT1 | -0.063 | -0.167 |
|

|
| ONECUT2_ONECUT3 | -0.216 | -0.443 |
|
|
| OSR1_OSR2 | -0.221 | -0.437 |
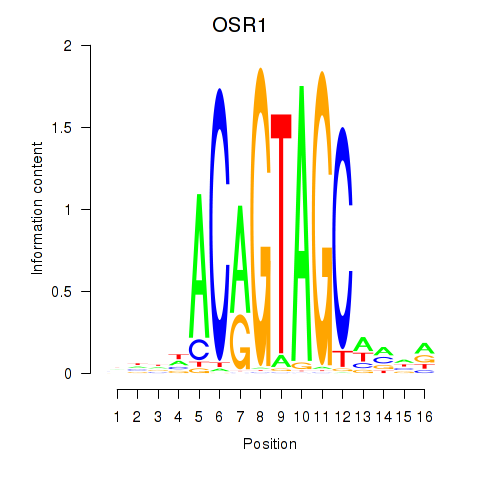
|
|
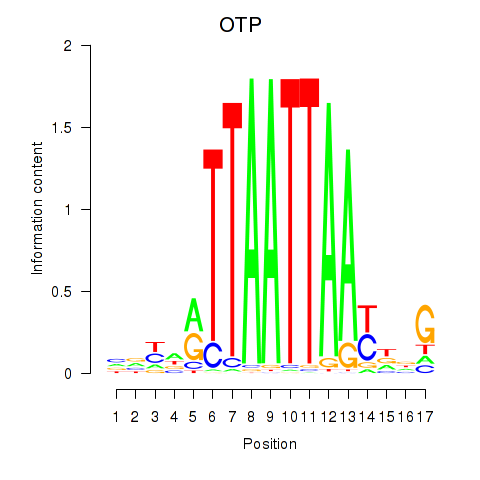
| OTP_PHOX2B_LHX1_LMX1A_LHX5_HOXC4 | 0.148 | 0.291 |
|
|
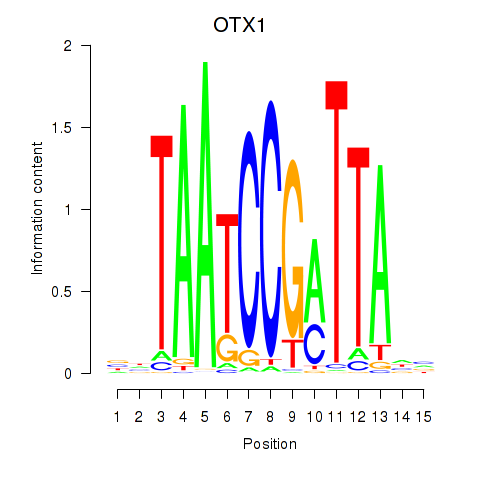
| OTX1 | 0.320 | 0.748 |
|
|
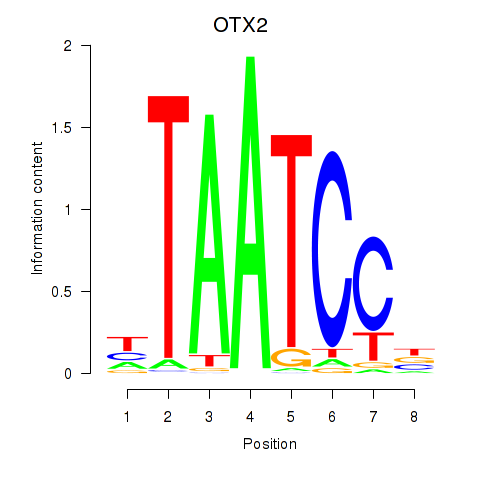
| OTX2_CRX | -0.268 | -0.556 |
|
|
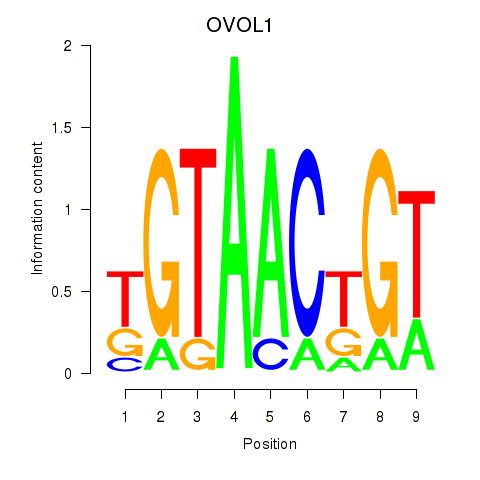
| OVOL1 | 0.835 | 0.573 |
|
|
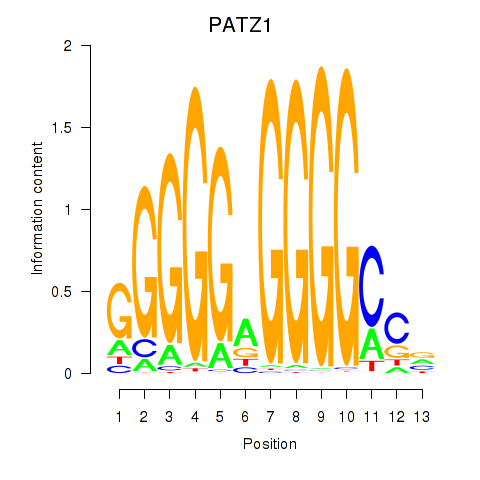
| PATZ1_KLF4 | -0.003 | -0.070 |
|
|
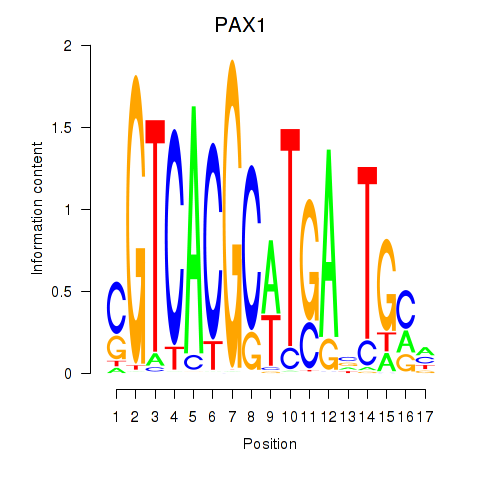
| PAX1_PAX9 | 0.324 | 0.458 |
|
|
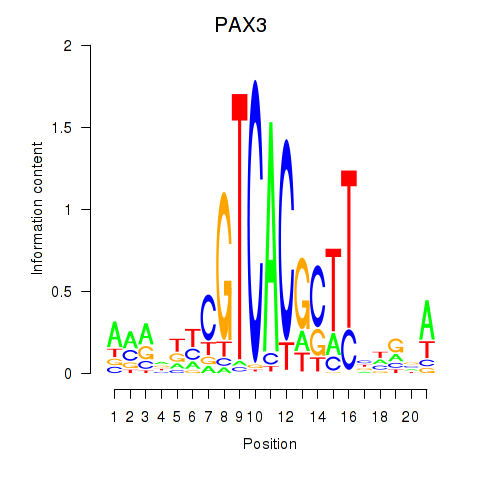
| PAX3 | 0.026 | 0.055 |
|
|
| PAX4 | 0.027 | 0.072 |
|
|
| PAX5 | 0.020 | 0.288 |
|
|
| PAX6 | -0.156 | -0.419 |
|
|
| PAX7_NOBOX | 0.073 | 0.199 |
|
|
| PAX8 | -0.259 | -0.638 |
|
|
| PBX1 | -0.059 | -0.147 |
|

|
| PBX2 | 0.285 | 0.295 |
|
|
| PBX3 | 0.069 | 0.309 |
|
|
| PDX1 | 0.094 | 0.185 |
|

|
| PGR | 0.024 | 0.071 |
|
|
| PITX1 | 0.155 | 1.499 |
|
|
| PITX2 | -0.187 | -0.194 |
|
|
| PITX3 | 0.187 | 1.368 |
|
|
| PKNOX1_TGIF2 | -0.245 | -0.676 |
|

|
| PKNOX2 | 0.093 | 0.154 |
|
|
| PLAG1 | -0.199 | -0.399 |
|
|
| PLAGL1 | -0.007 | -0.122 |
|
|
| POU1F1 | 0.219 | 0.501 |
|

|
| POU2F1 | 0.073 | 0.165 |
|
|
| POU2F2_POU3F1 | -0.040 | -0.135 |
|
|
| POU3F2 | 0.040 | 0.341 |
|
|
| POU3F3_POU3F4 | -0.230 | -0.683 |
|
|
| POU4F1_POU4F3 | 0.135 | 0.347 |
|
|
| POU4F2 | -0.079 | -0.111 |
|
|
| POU5F1_POU2F3 | -0.209 | -0.994 |
|
|
| POU6F1 | 0.195 | 0.287 |
|

|
| POU6F2 | 0.005 | 0.009 |
|
|
| PPARA | 0.125 | 0.440 |
|
|
| PPARD | -0.375 | -0.609 |
|
|
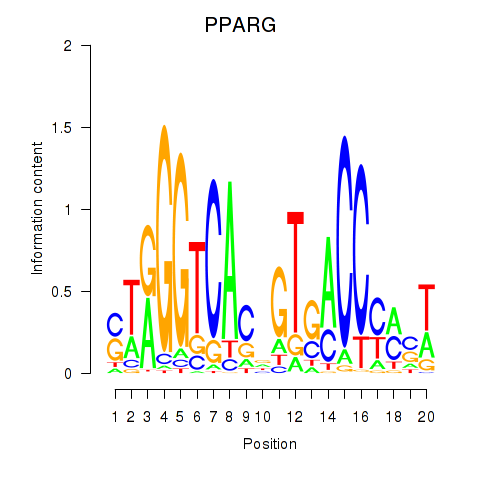
| PPARG | -0.155 | -0.554 |
|
|
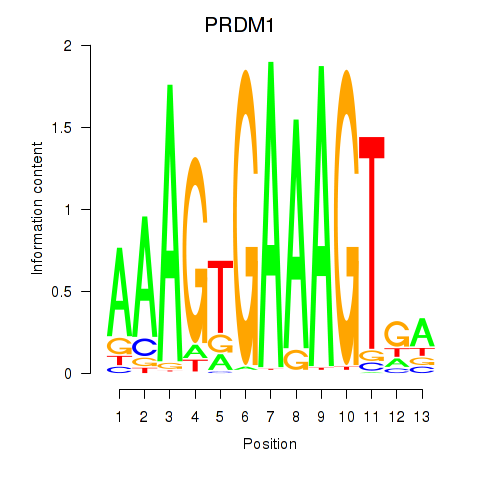
| PRDM1 | -0.146 | -0.366 |
|
|
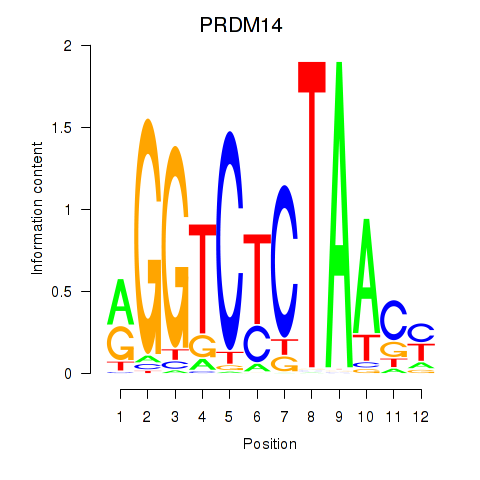
| PRDM14 | -0.100 | -0.125 |
|
|
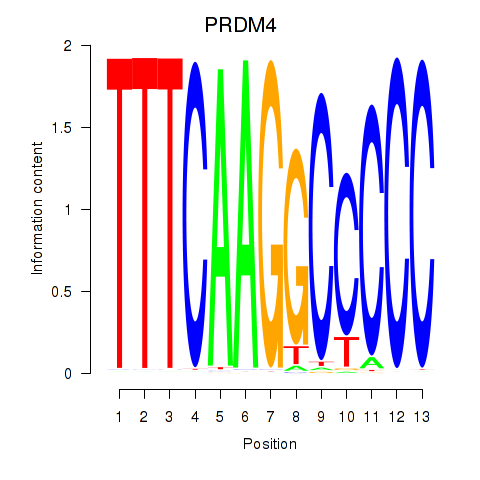
| PRDM4 | -0.183 | -0.253 |
|
|
| PROX1 | 0.113 | 0.303 |
|
|
| PRRX1_ALX4_PHOX2A | 0.718 | 1.174 |
|
|
| PTF1A | -0.009 | -0.018 |
|
|
| RAD21_SMC3 | 0.192 | 0.713 |
|
|
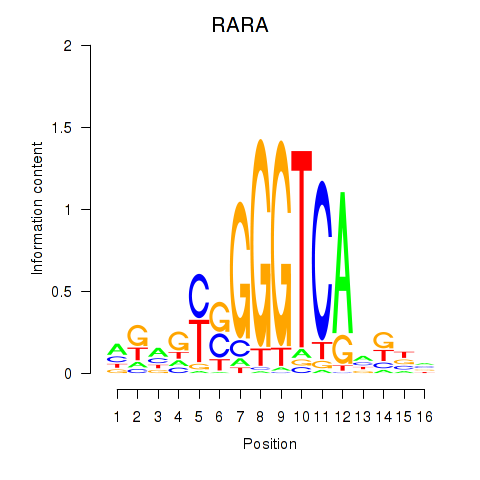
| RARA | -0.190 | -0.624 |
|
|
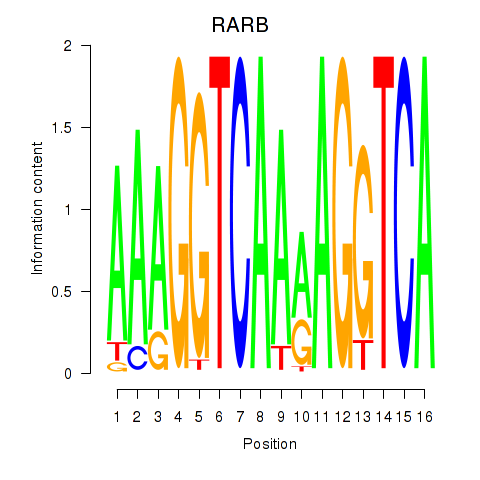
| RARB | -0.244 | -0.633 |
|
|
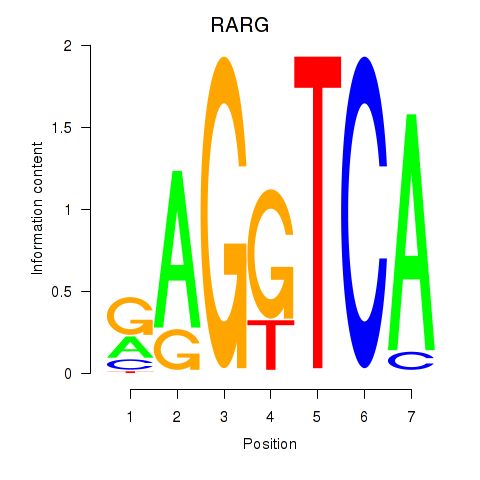
| RARG | 0.029 | 0.164 |
|
|
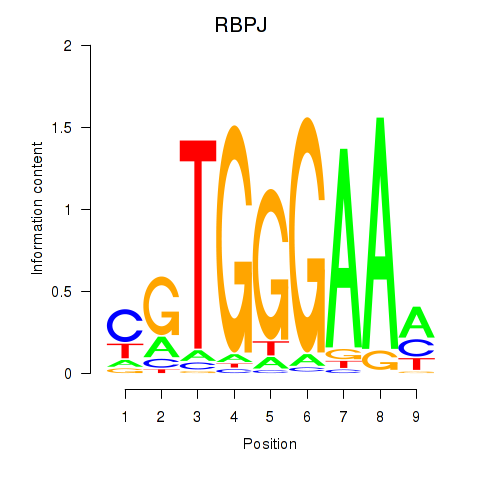
| RBPJ | -0.062 | -0.137 |
|
|
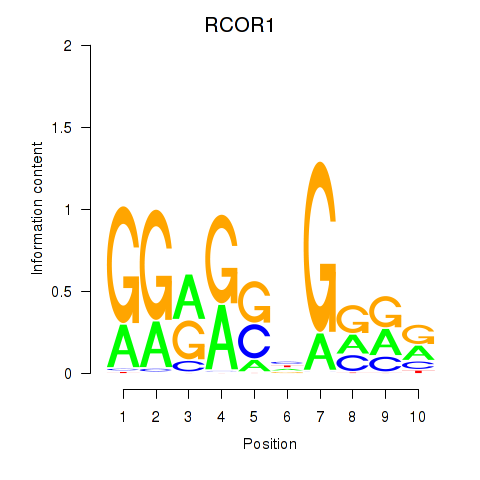
| RCOR1_MTA3 | 0.018 | 0.667 |
|
|
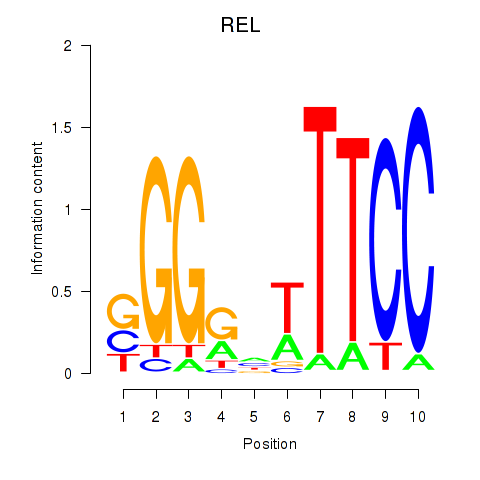
| REL | 0.063 | 0.161 |
|
|
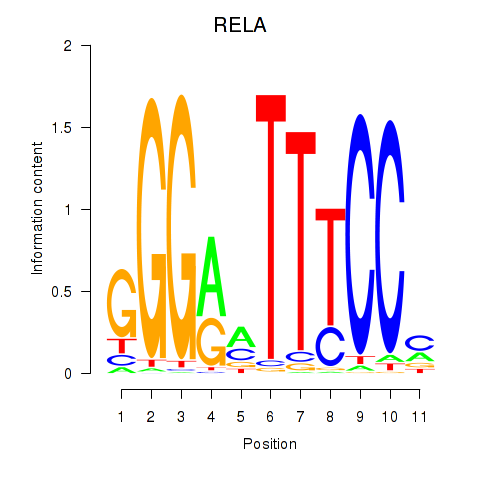
| RELA | -0.150 | -0.362 |
|
|
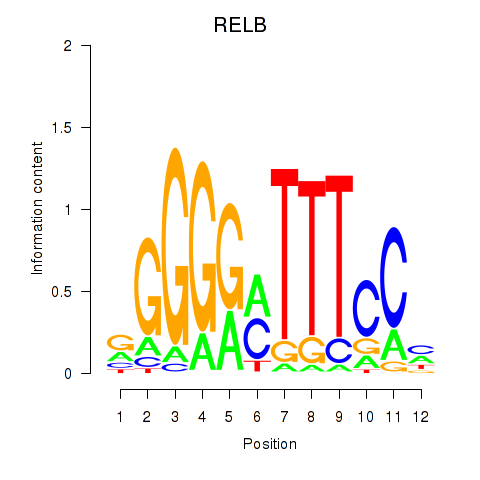
| RELB | -0.205 | -0.588 |
|
|
| REST | -0.649 | -1.344 |
|
|
| RFX3_RFX2 | -0.044 | -0.297 |
|
|
| RFX5 | -0.040 | -0.058 |
|
|
| RFX7_RFX4_RFX1 | -0.118 | -0.535 |
|
|
| RHOXF1 | -0.208 | -1.211 |
|
|
| RORA | 0.189 | 0.379 |
|
|
| RORC | -0.629 | -0.787 |
|
|
| RREB1 | -0.242 | -0.752 |
|
|
| RUNX1_RUNX2 | -0.104 | -0.605 |
|
|
| RUNX3_BCL11A | -0.140 | -0.435 |
|

|
| RXRA_NR2F6_NR2C2 | 0.076 | 0.266 |
|
|
| RXRG | -0.061 | -0.299 |
|

|
| SCRT1_SCRT2 | 0.092 | 0.257 |
|
|
| SHOX | -0.156 | -0.317 |
|
|
| SHOX2_HOXC5 | 0.005 | 0.023 |
|
|
| SIN3A_CHD1 | -0.010 | -0.152 |
|
|
| SIX1_SIX3_SIX2 | 0.048 | 0.131 |
|
|
| SIX4 | -0.130 | -0.201 |
|

|
| SIX5_SMARCC2_HCFC1 | 0.110 | 1.498 |
|
|
| SIX6 | 0.207 | 0.397 |
|
|
| SMAD1 | -0.119 | -1.685 |
|
|
| SMAD2 | -0.288 | -0.674 |
|
|
| SMAD3 | -0.480 | -0.986 |
|
|
| SMAD4 | 0.048 | 0.290 |
|
|
| SOX1 | -0.341 | -0.624 |
|
|
| SOX10_SOX15 | 0.283 | 0.700 |
|
|
| SOX11 | -0.271 | -0.519 |
|
|
| SOX13_SOX12 | -0.124 | -0.558 |
|
|
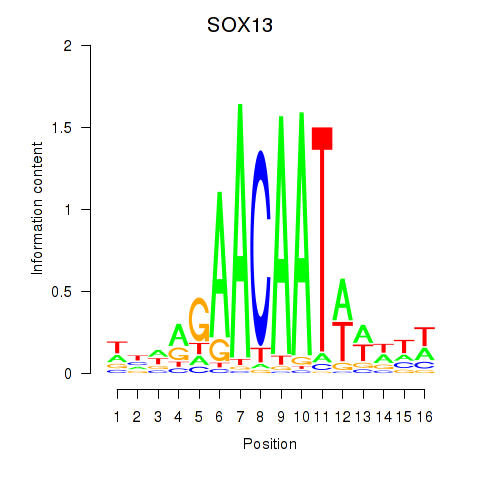
| SOX14 | 0.076 | 0.180 |
|
|
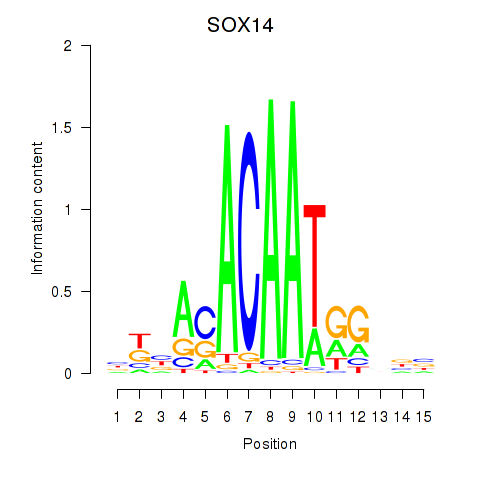
| SOX17 | 0.154 | 0.511 |
|
|
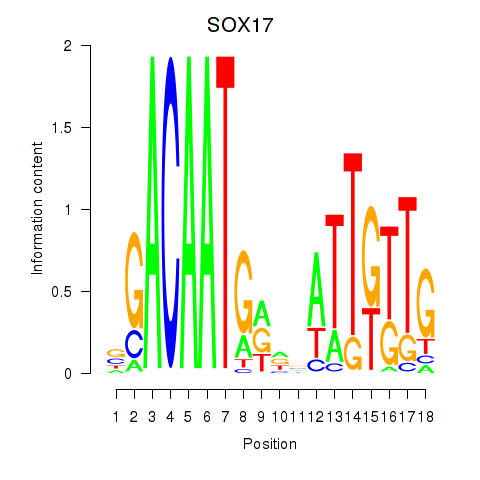
| SOX18 | -0.119 | -0.279 |
|
|
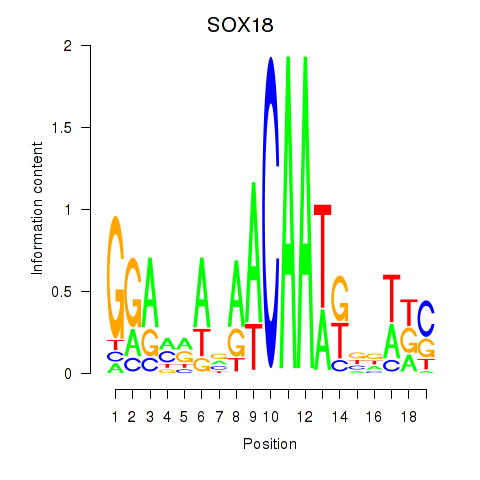
| SOX21 | -0.031 | -0.044 |
|
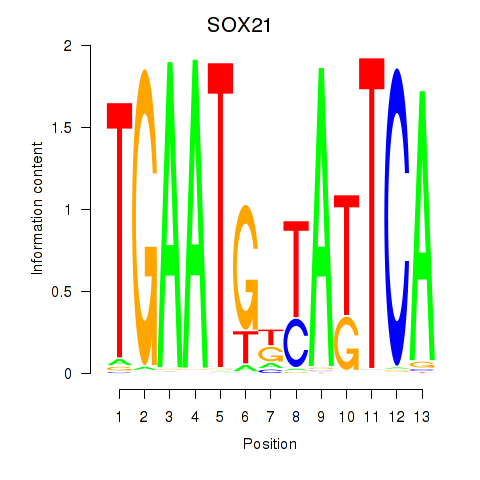
|
| SOX30 | 0.384 | 0.591 |
|

|
| SOX3_SOX2 | 0.018 | 0.103 |
|
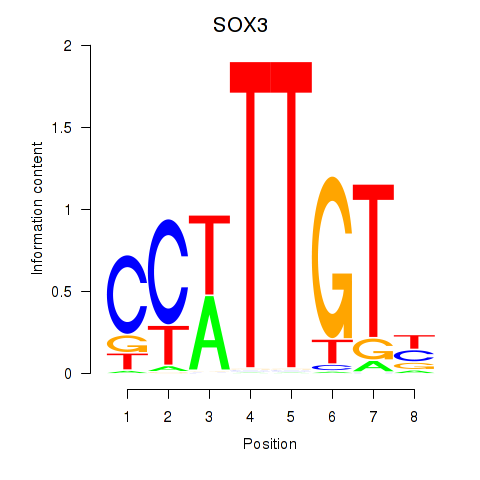
|
| SOX4 | -0.119 | -0.276 |
|
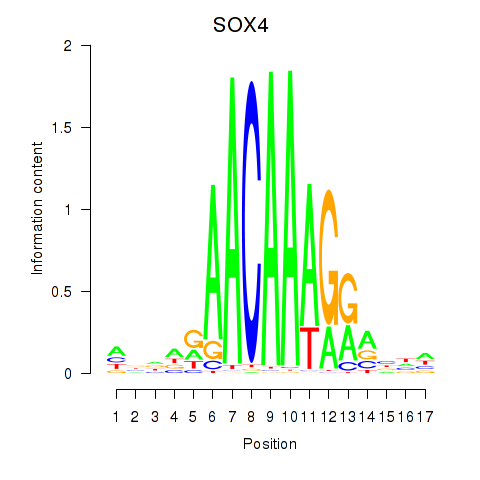
|
| SOX5 | 0.112 | 0.278 |
|
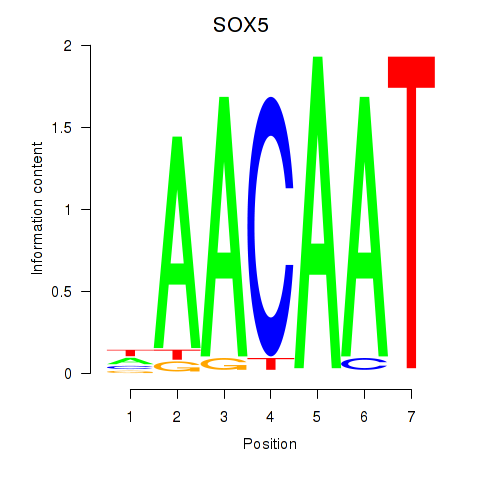
|
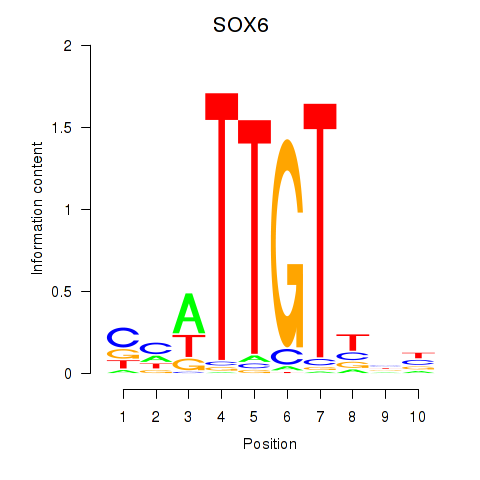
| SOX6 | 0.059 | 0.142 |
|
|
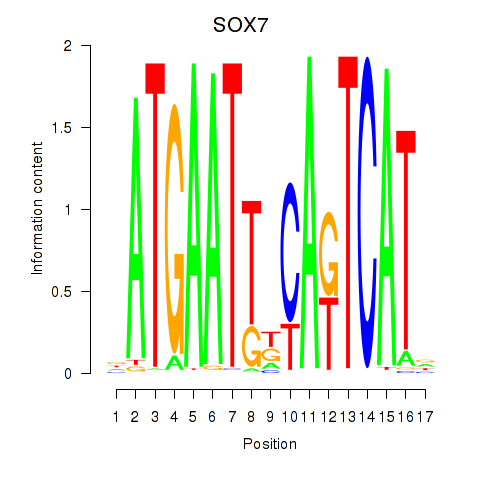
| SOX7 | 0.230 | 0.305 |
|
|
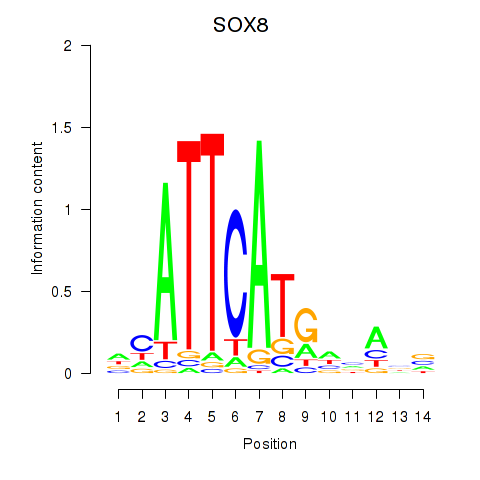
| SOX8 | 0.079 | 0.212 |
|
|
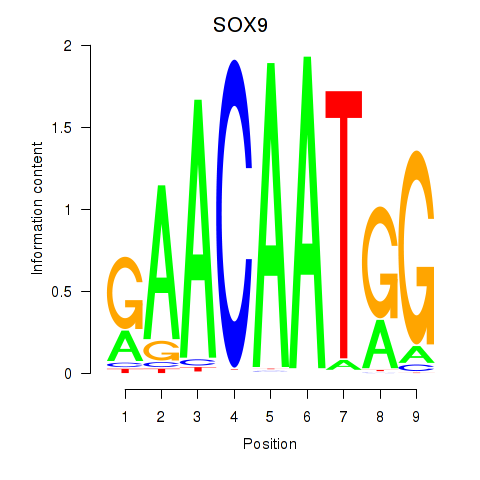
| SOX9 | 0.031 | 0.116 |
|
|
| SP1 | 0.023 | 0.383 |
|

|
| SP100 | 0.505 | 2.017 |
|
|
| SP3 | 0.037 | 0.908 |
|

|
| SP4_PML | -0.047 | -1.209 |
|
|
| SPDEF | 0.129 | 0.354 |
|
|
| SPI1 | 0.002 | 0.012 |
|
|
| SPIB | 0.301 | 1.101 |
|
|
| SPIC | -0.087 | -0.440 |
|
|
| SREBF1_TFE3 | -0.126 | -0.524 |
|
|
| SREBF2 | 0.563 | 0.836 |
|
|
| SRF | 0.215 | 0.542 |
|
|
| SRY | -0.104 | -0.258 |
|
|
| STAT1_STAT3_BCL6 | -0.046 | -0.169 |
|
|
| STAT4 | 0.179 | 0.384 |
|
|
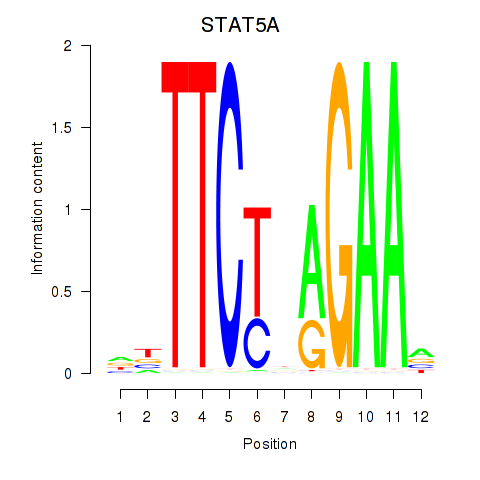
| STAT5A | -0.183 | -1.055 |
|
|
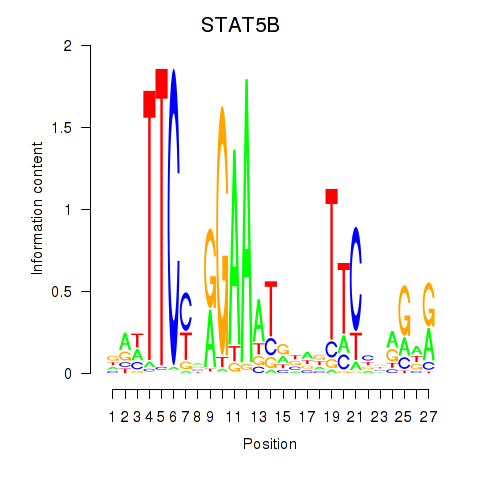
| STAT5B | -0.138 | -0.384 |
|
|
| STAT6 | -0.374 | -0.846 |
|

|
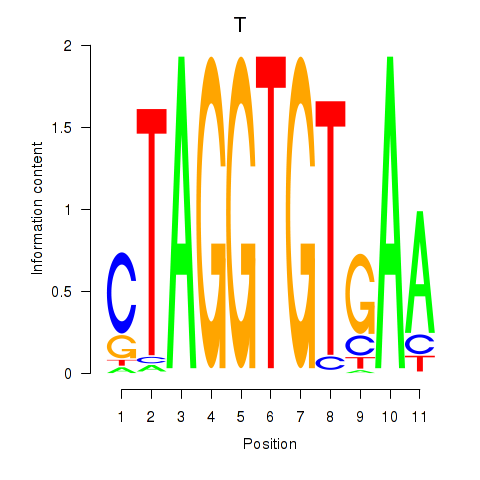
| T | -0.029 | -0.058 |
|
|
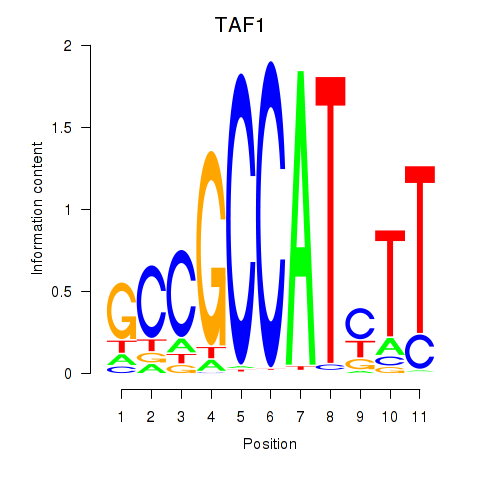
| TAF1 | 0.155 | 0.964 |
|
|
| TAL1 | -0.060 | -0.759 |
|
|
| TBP | 0.186 | 0.762 |
|
|
| TBX1 | 0.021 | 0.145 |
|
|
| TBX15_MGA | -0.045 | -0.140 |
|
|
| TBX19 | -0.087 | -0.199 |
|
|
| TBX2 | -0.030 | -0.087 |
|
|
| TBX20 | 0.255 | 0.403 |
|
|
| TBX21_TBR1 | -0.041 | -0.112 |
|
|
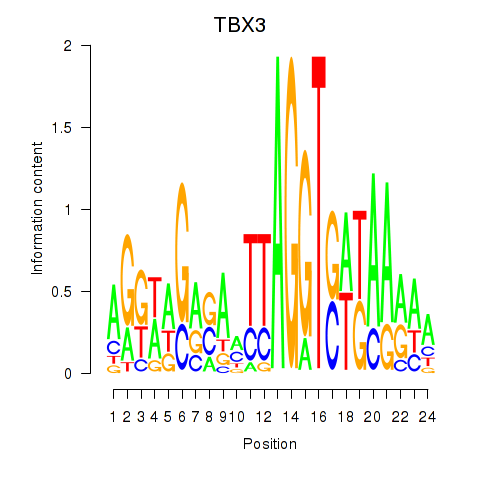
| TBX3 | 0.000 | 0.000 |
|
|
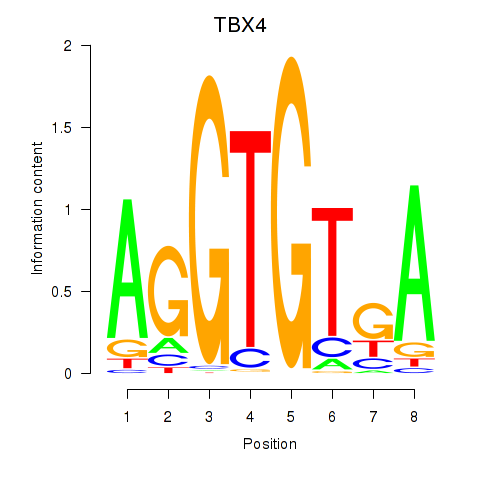
| TBX4 | 0.145 | 0.223 |
|
|
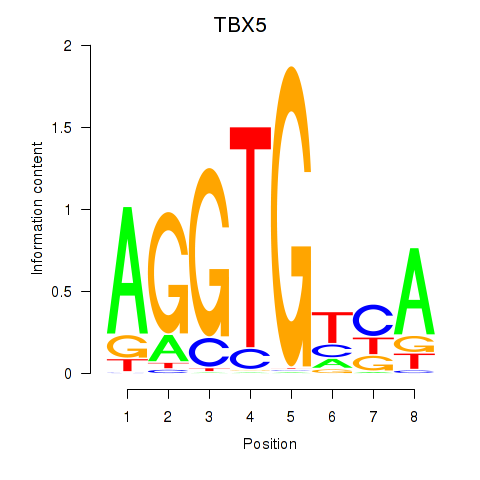
| TBX5 | -0.033 | -0.107 |
|
|
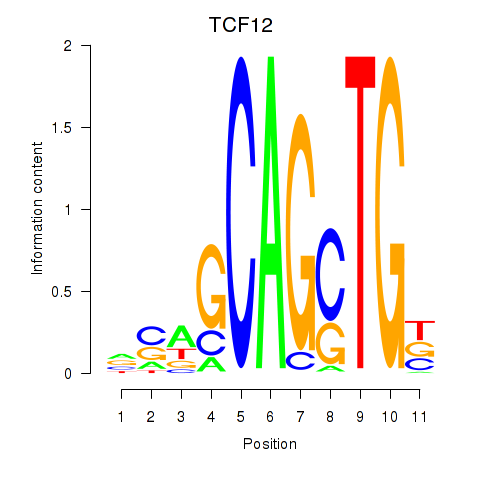
| TCF12_ASCL2 | -0.006 | -0.039 |
|
|
| TCF21 | -0.095 | -0.315 |
|
|
| TCF3_MYOG | -0.023 | -0.096 |
|
|
| TCF7 | 0.239 | 0.705 |
|

|
| TCF7L1 | -0.044 | -0.096 |
|
|
| TCF7L2 | 0.067 | 0.139 |
|
|
| TEAD3_TEAD1 | 0.012 | 0.049 |
|
|
| TEAD4 | -0.248 | -0.646 |
|
|
| TFAP2B | -0.109 | -0.439 |
|
|
| TFAP2C | -0.088 | -0.859 |
|
|
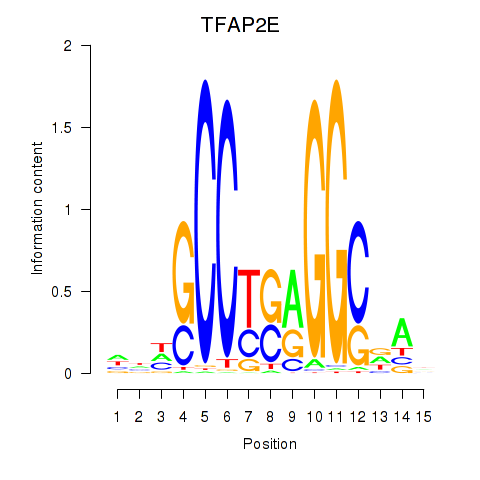
| TFAP2E | -0.162 | -0.286 |
|
|
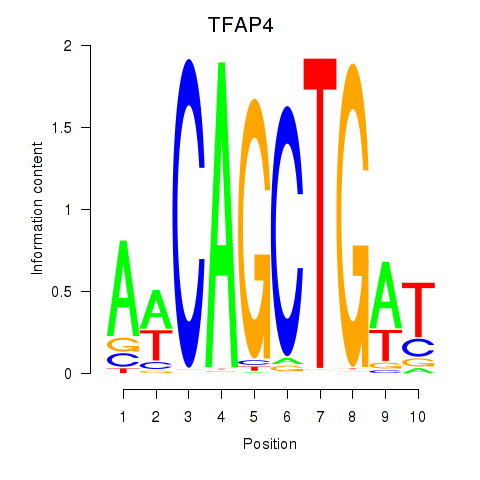
| TFAP4_MSC | 0.096 | 0.332 |
|
|
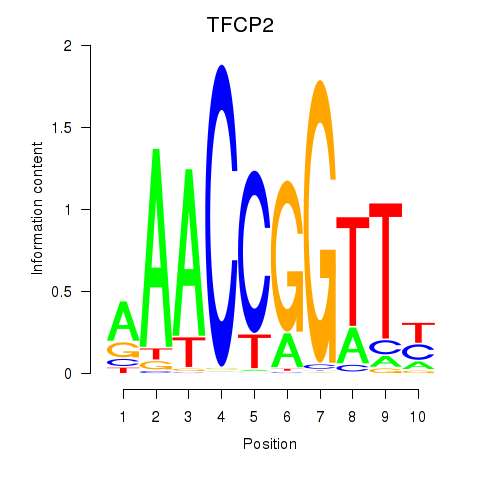
| TFCP2 | 0.220 | 0.670 |
|
|
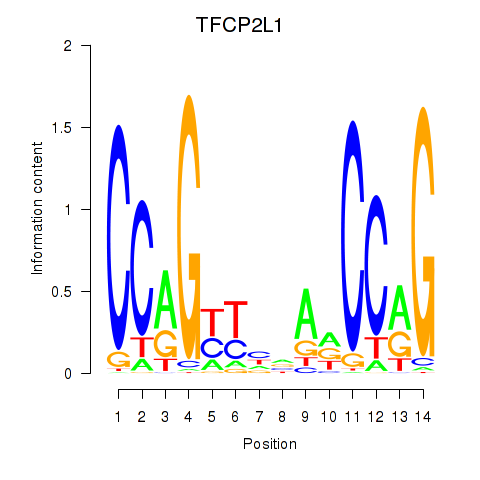
| TFCP2L1 | -0.000 | -0.001 |
|
|
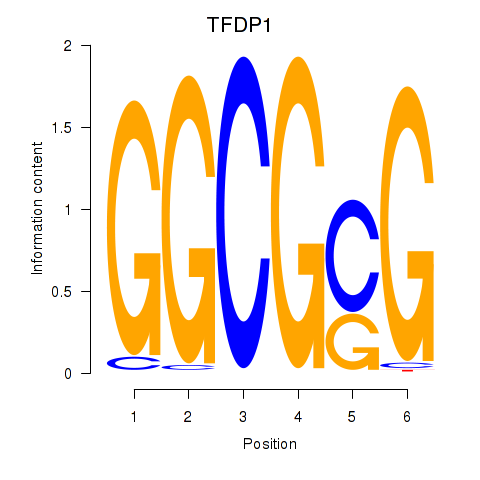
| TFDP1 | 0.050 | 0.467 |
|
|
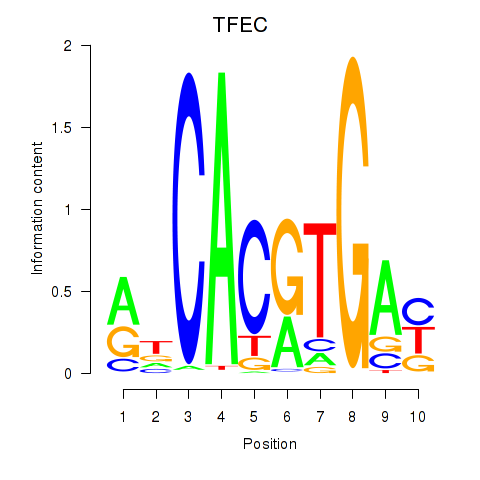
| TFEC_MITF_ARNTL_BHLHE41 | 0.174 | 0.665 |
|
|
| TGIF1 | -0.097 | -0.357 |
|

|
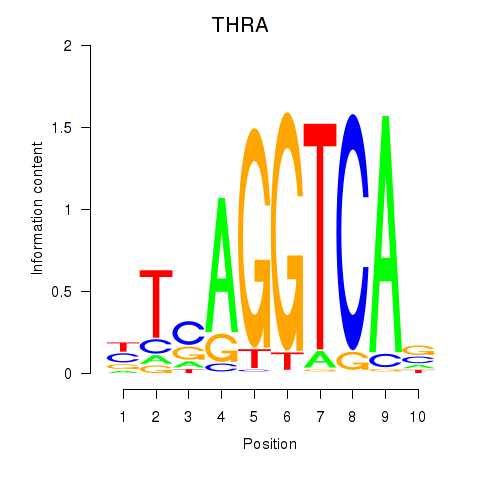
| THRA_RXRB | -0.075 | -0.207 |
|
|
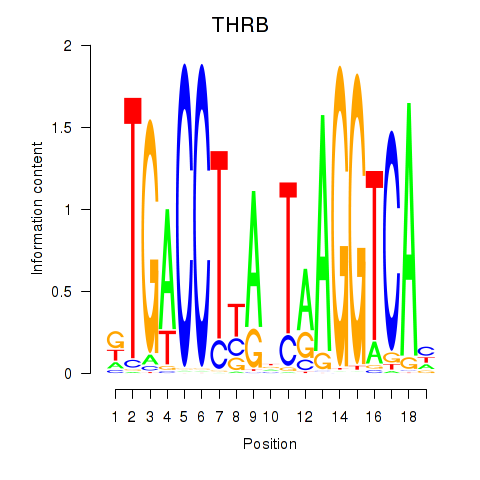
| THRB | 0.174 | 0.209 |
|
|
| TLX1_NFIC | 0.097 | 0.363 |
|
|
| TLX2 | 0.095 | 0.380 |
|
|
| TP53 | -0.060 | -0.182 |
|
|
| TP63 | 0.266 | 0.826 |
|
|
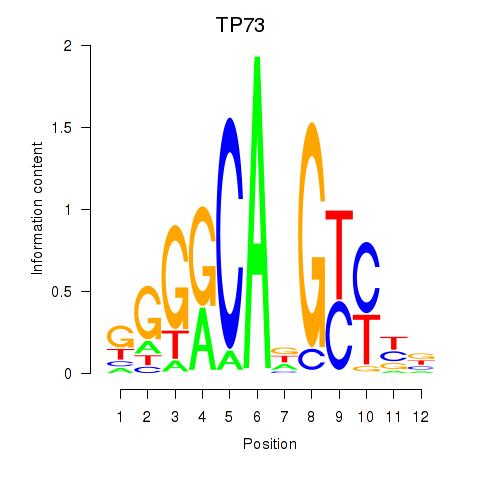
| TP73 | -0.118 | -0.187 |
|
|
| TWIST1_SNAI1 | -0.091 | -0.424 |
|

|
| UAAGACG | 1.107 | 0.489 |
|

|
| UAAGACU | -0.642 | -0.266 |
|

|
| UAAGGCA | 0.140 | 0.400 |
|

|
| UAAUGCU | -0.351 | -0.438 |
|

|
| UACAGUA | 0.043 | 0.052 |
|

|
| UAGUGUU | 0.279 | 0.500 |
|
|
| UAUUGCU | 0.016 | 0.053 |
|

|
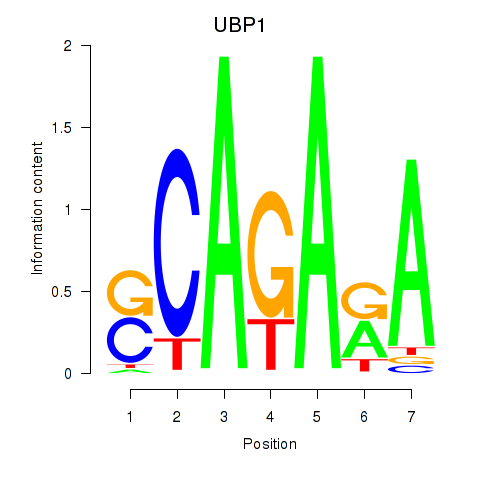
| UBP1 | 0.048 | 0.063 |
|
|
| UCACAGU | -0.219 | -0.465 |
|

|
| UCACAUU | -0.316 | -0.567 |
|

|
| UCCAGUU | 0.287 | 0.711 |
|

|
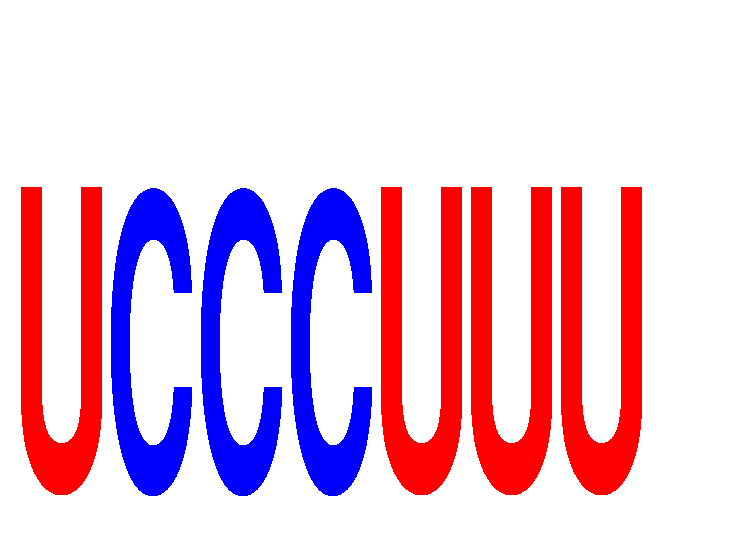
| UCCCUUU | -0.066 | -0.065 |
|
|
| UGAAAUG | 0.306 | 0.370 |
|
|
| UGACAUC | 0.567 | 0.292 |
|

|
| UGACCUA | 0.321 | 0.145 |
|

|
| UGCAGUC | 0.174 | 0.158 |
|

|
| UGCAUAG | -0.014 | -0.041 |
|

|
| UGCAUUG | 0.210 | 0.188 |
|

|
| UGGCACU | 0.240 | 0.472 |
|
|
| UGGUCCC | 0.028 | 0.059 |
|

|
| UGUGCGU | 1.360 | 0.352 |
|

|
| UGUGCUU | -0.207 | -0.614 |
|

|
| UUGGCAA | 0.225 | 0.408 |
|

|
| UUGGCAC | -0.318 | -0.566 |
|

|
| UUGGUCC | 0.193 | 0.428 |
|

|
| UUGUUCG | -0.343 | -0.324 |
|

|
| UUUUUGC | 0.510 | 0.128 |
|

|
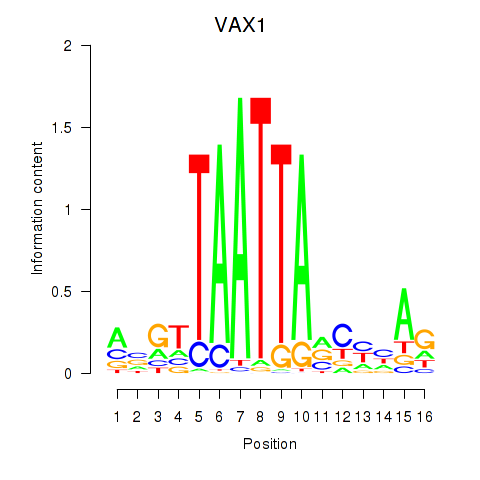
| VAX1_GSX2 | -0.555 | -0.978 |
|
|
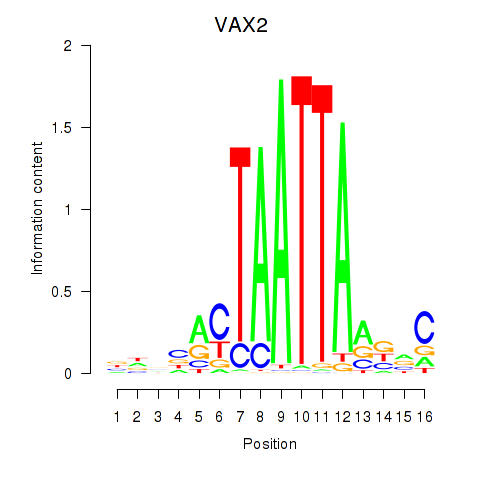
| VAX2_RHOXF2 | 0.459 | 0.572 |
|
|
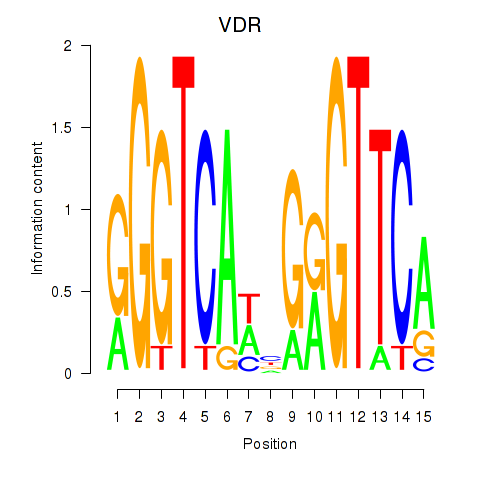
| VDR | 0.087 | 0.259 |
|
|
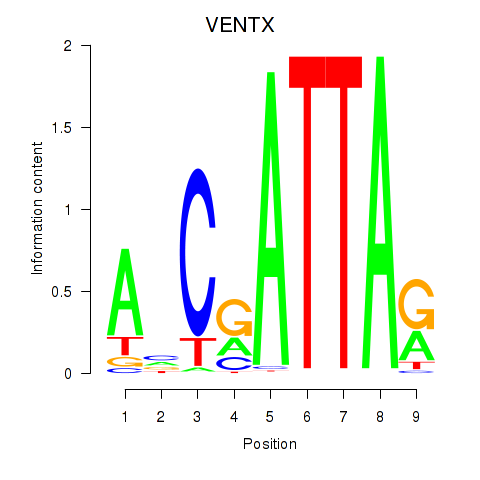
| VENTX | 0.205 | 0.364 |
|
|
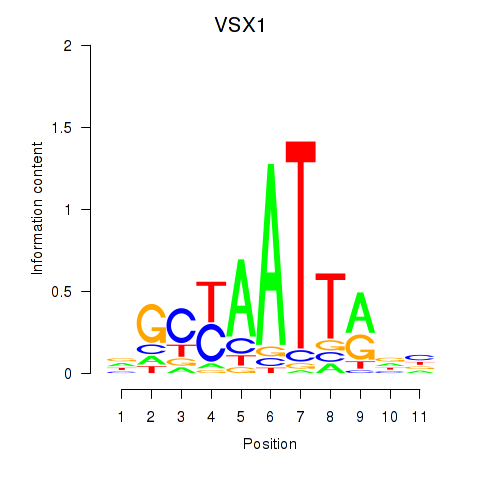
| VSX1 | 0.146 | 0.389 |
|
|
| WRNIP1 | -0.071 | -1.383 |
|
|
| WT1_MTF1_ZBTB7B | -0.032 | -0.545 |
|
|
| XBP1 | 0.103 | 0.246 |
|
|
| YBX1_FOS_NFYC_NFYA_NFYB_CEBPZ | 0.060 | 1.259 |
|
|
| YY1_YY2 | 0.235 | 1.315 |
|
|
| ZBED1 | -0.868 | -0.638 |
|
|
| ZBTB12 | 0.235 | 0.426 |
|
|
| ZBTB14 | -0.079 | -0.577 |
|
|
| ZBTB16 | 0.000 | 0.001 |
|
|
| ZBTB18 | -0.355 | -0.815 |
|
|
| ZBTB3 | -0.055 | -0.089 |
|

|
| ZBTB33_CHD2 | 0.306 | 2.273 |
|
|
| ZBTB4 | -0.012 | -0.027 |
|
|
| ZBTB49 | -0.171 | -0.272 |
|
|
| ZBTB6 | -0.053 | -0.273 |
|
|
| ZBTB7A_ZBTB7C | 0.061 | 0.147 |
|

|
| ZEB1 | -0.066 | -0.712 |
|
|
| ZFHX3 | -0.137 | -0.176 |
|

|
| ZFX | 0.114 | 0.972 |
|
|
| ZIC1 | -0.325 | -0.706 |
|
|
| ZIC2_GLI1 | -0.184 | -0.483 |
|
|
| ZIC3_ZIC4 | -0.136 | -0.529 |
|
|
| ZKSCAN1 | 0.124 | 0.265 |
|
|
| ZKSCAN3 | 0.201 | 0.267 |
|
|
| ZNF143 | -0.001 | -0.006 |
|
|
| ZNF148 | 0.005 | 0.021 |
|
|
| ZNF232 | 0.415 | 0.546 |
|
|
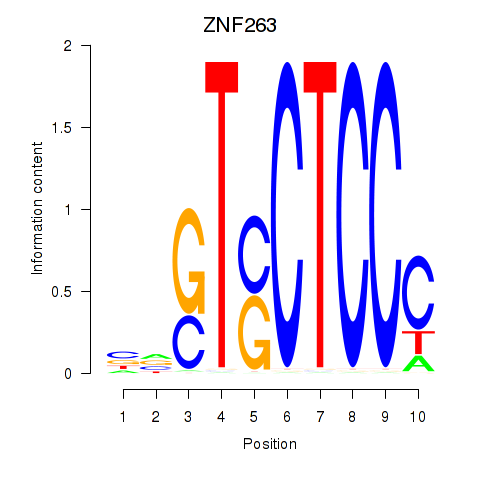
| ZNF263 | -0.184 | -1.252 |
|
|
| ZNF274 | 0.203 | 0.225 |
|

|
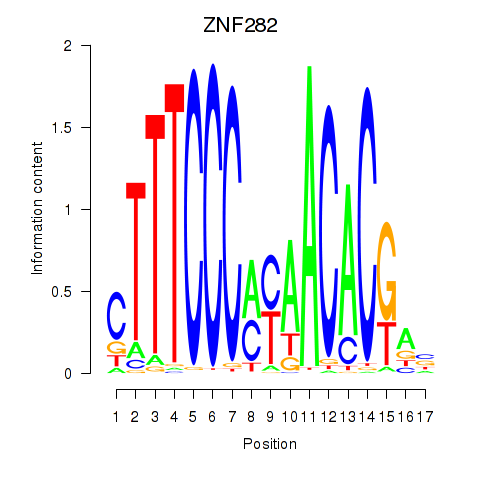
| ZNF282 | 0.163 | 0.308 |
|
|
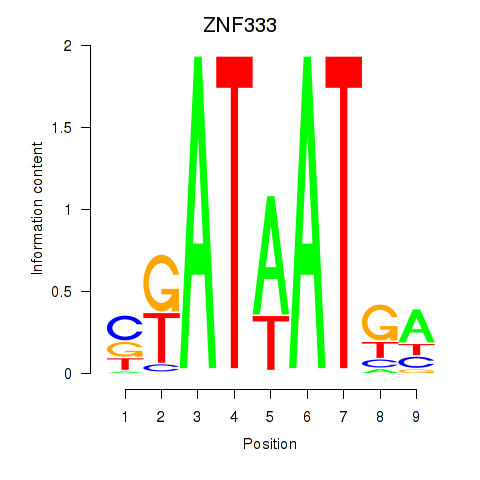
| ZNF333 | -0.172 | -0.294 |
|
|
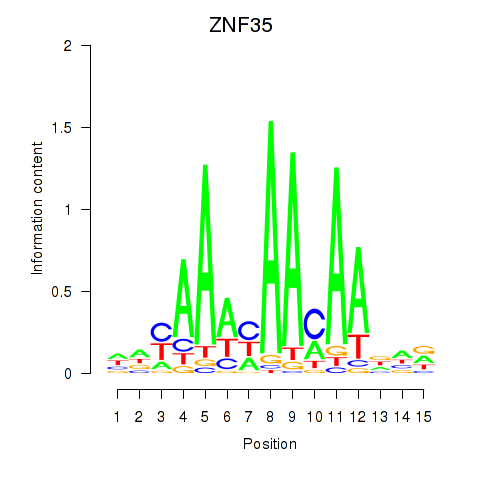
| ZNF35 | -0.142 | -0.141 |
|
|
| ZNF350 | -0.000 | -0.000 |
|
|
| ZNF384 | 0.051 | 0.494 |
|
|
| ZNF410 | 0.279 | 0.377 |
|
|
| ZNF423 | -0.077 | -0.150 |
|
|
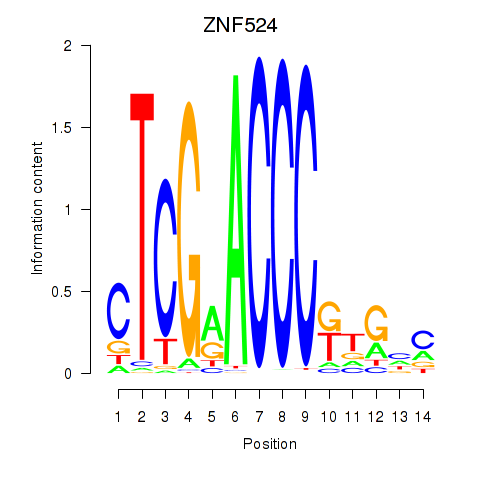
| ZNF524 | 0.017 | 0.116 |
|
|
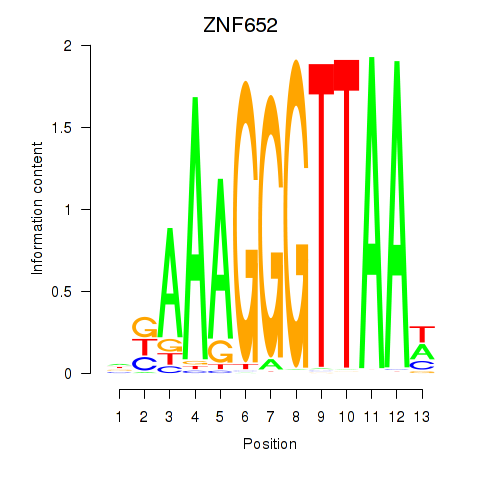
| ZNF652 | -0.068 | -0.253 |
|
|
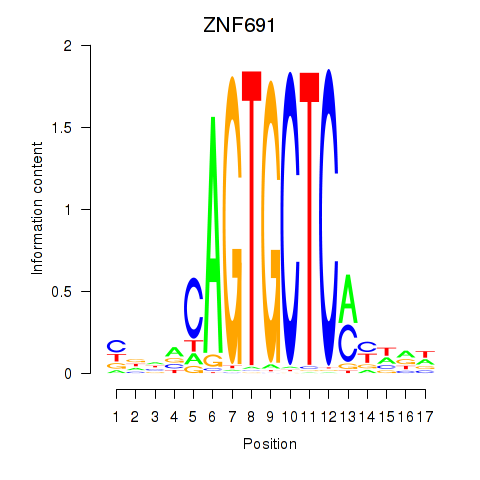
| ZNF691 | -0.259 | -0.461 |
|
|
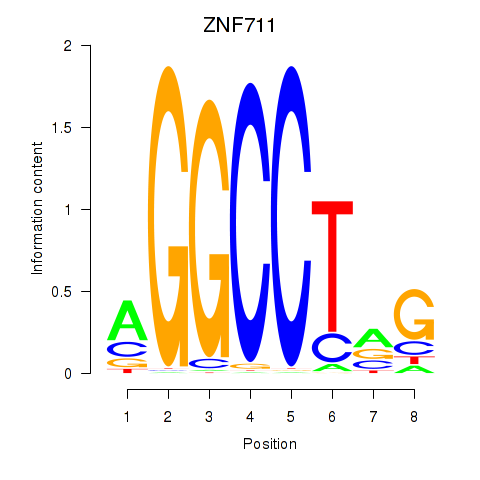
| ZNF711_TFAP2A_TFAP2D | 0.049 | 1.262 |
|
|
| ZNF740_ZNF219 | 0.133 | 0.534 |
|
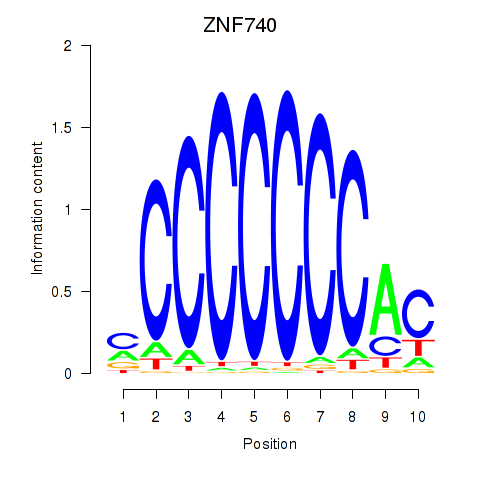
|
| ZNF784 | -0.102 | -0.312 |
|
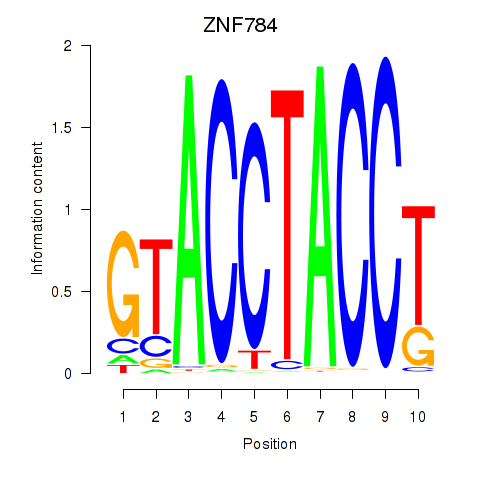
|
| ZNF8 | 0.259 | 0.339 |
|
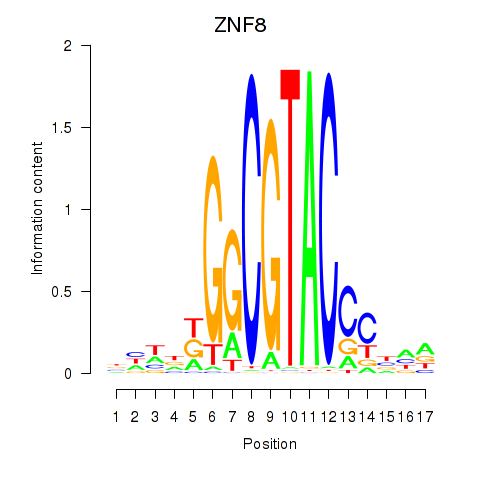
|
| ZSCAN4 | 0.004 | 0.009 |
|
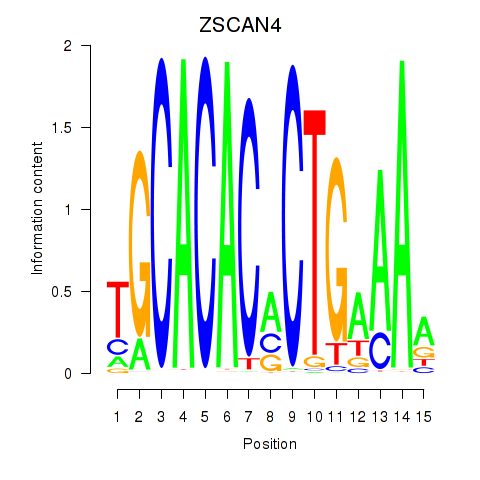
|