Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
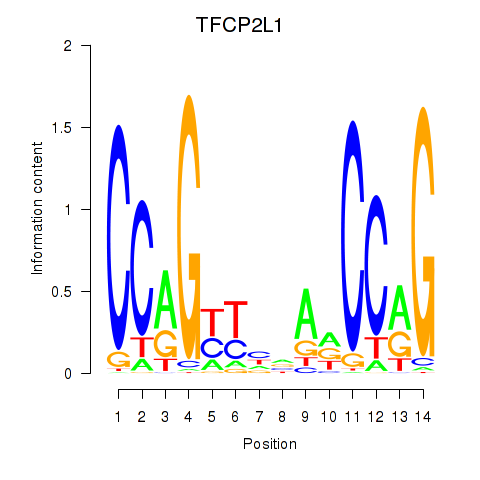
Results for TFCP2L1
Z-value: 0.03
Transcription factors associated with TFCP2L1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFCP2L1
|
ENSG00000115112.7 | transcription factor CP2 like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TFCP2L1 | hg19_v2_chr2_-_122042770_122042785 | 0.02 | 7.5e-01 | Click! |
Activity profile of TFCP2L1 motif
Sorted Z-values of TFCP2L1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_1071203 | 15.88 |
ENST00000543365.1
|
HMHA1
|
histocompatibility (minor) HA-1 |
| chr2_-_136875712 | 13.84 |
ENST00000241393.3
|
CXCR4
|
chemokine (C-X-C motif) receptor 4 |
| chr6_-_31550192 | 10.18 |
ENST00000429299.2
ENST00000446745.2 |
LTB
|
lymphotoxin beta (TNF superfamily, member 3) |
| chr20_-_43280361 | 9.72 |
ENST00000372874.4
|
ADA
|
adenosine deaminase |
| chr5_-_131826457 | 8.72 |
ENST00000437654.1
ENST00000245414.4 |
IRF1
|
interferon regulatory factor 1 |
| chr20_-_43280325 | 8.40 |
ENST00000537820.1
|
ADA
|
adenosine deaminase |
| chr1_-_111743285 | 7.42 |
ENST00000357640.4
|
DENND2D
|
DENN/MADD domain containing 2D |
| chr1_-_161600990 | 7.39 |
ENST00000531221.1
|
FCGR3B
|
Fc fragment of IgG, low affinity IIIb, receptor (CD16b) |
| chr20_-_62710832 | 7.24 |
ENST00000395042.1
|
RGS19
|
regulator of G-protein signaling 19 |
| chr1_-_161600942 | 7.06 |
ENST00000421702.2
|
FCGR3B
|
Fc fragment of IgG, low affinity IIIb, receptor (CD16b) |
| chr1_-_153538011 | 6.84 |
ENST00000368707.4
|
S100A2
|
S100 calcium binding protein A2 |
| chr6_+_31555045 | 6.79 |
ENST00000396101.3
ENST00000490742.1 |
LST1
|
leukocyte specific transcript 1 |
| chr1_-_161600822 | 6.78 |
ENST00000534776.1
ENST00000540048.1 |
FCGR3B
FCGR3A
|
Fc fragment of IgG, low affinity IIIb, receptor (CD16b) Fc fragment of IgG, low affinity IIIa, receptor (CD16a) |
| chr14_+_64970662 | 6.74 |
ENST00000556965.1
ENST00000554015.1 |
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr22_-_37545972 | 6.32 |
ENST00000216223.5
|
IL2RB
|
interleukin 2 receptor, beta |
| chrX_-_106960285 | 6.28 |
ENST00000503515.1
ENST00000372397.2 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr15_-_62352570 | 6.16 |
ENST00000261517.5
ENST00000395896.4 ENST00000395898.3 |
VPS13C
|
vacuolar protein sorting 13 homolog C (S. cerevisiae) |
| chr1_+_27189631 | 6.15 |
ENST00000339276.4
|
SFN
|
stratifin |
| chr17_-_76124711 | 5.83 |
ENST00000306591.7
ENST00000590602.1 |
TMC6
|
transmembrane channel-like 6 |
| chr6_+_31554962 | 5.45 |
ENST00000376092.3
ENST00000376086.3 ENST00000303757.8 ENST00000376093.2 ENST00000376102.3 |
LST1
|
leukocyte specific transcript 1 |
| chr10_-_121296045 | 5.41 |
ENST00000392865.1
|
RGS10
|
regulator of G-protein signaling 10 |
| chr4_-_25865159 | 5.40 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr1_-_161519579 | 5.32 |
ENST00000426740.1
|
FCGR3A
|
Fc fragment of IgG, low affinity IIIa, receptor (CD16a) |
| chr17_+_75372165 | 5.06 |
ENST00000427674.2
|
SEPT9
|
septin 9 |
| chr1_-_161519682 | 5.01 |
ENST00000367969.3
ENST00000443193.1 |
FCGR3A
|
Fc fragment of IgG, low affinity IIIa, receptor (CD16a) |
| chr2_+_98330009 | 4.95 |
ENST00000264972.5
|
ZAP70
|
zeta-chain (TCR) associated protein kinase 70kDa |
| chr17_-_76124812 | 4.82 |
ENST00000592063.1
ENST00000589271.1 ENST00000322933.4 ENST00000589553.1 |
TMC6
|
transmembrane channel-like 6 |
| chr14_-_107170409 | 4.76 |
ENST00000390633.2
|
IGHV1-69
|
immunoglobulin heavy variable 1-69 |
| chr1_-_153538292 | 4.75 |
ENST00000497140.1
ENST00000368708.3 |
S100A2
|
S100 calcium binding protein A2 |
| chr11_+_128562372 | 4.62 |
ENST00000344954.6
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr6_-_32731299 | 4.53 |
ENST00000435145.2
ENST00000437316.2 |
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2 |
| chr2_-_89385283 | 4.51 |
ENST00000390252.2
|
IGKV3-15
|
immunoglobulin kappa variable 3-15 |
| chr11_+_46383121 | 4.46 |
ENST00000454345.1
|
DGKZ
|
diacylglycerol kinase, zeta |
| chr16_-_86542455 | 4.32 |
ENST00000595886.1
ENST00000597578.1 ENST00000593604.1 |
FENDRR
|
FOXF1 adjacent non-coding developmental regulatory RNA |
| chr1_-_89458636 | 4.10 |
ENST00000370486.1
ENST00000399794.2 |
CCBL2
RBMXL1
|
cysteine conjugate-beta lyase 2 RNA binding motif protein, X-linked-like 1 |
| chr8_-_117778494 | 4.02 |
ENST00000276682.4
|
EIF3H
|
eukaryotic translation initiation factor 3, subunit H |
| chr17_+_16318909 | 4.02 |
ENST00000577397.1
|
TRPV2
|
transient receptor potential cation channel, subfamily V, member 2 |
| chr4_-_83351294 | 4.00 |
ENST00000502762.1
|
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr19_-_12721616 | 3.98 |
ENST00000311437.6
|
ZNF490
|
zinc finger protein 490 |
| chr14_-_64971288 | 3.91 |
ENST00000394715.1
|
ZBTB25
|
zinc finger and BTB domain containing 25 |
| chr12_+_56324756 | 3.87 |
ENST00000331886.5
ENST00000555090.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr12_+_56324933 | 3.86 |
ENST00000549629.1
ENST00000555218.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr17_+_75369167 | 3.86 |
ENST00000423034.2
|
SEPT9
|
septin 9 |
| chr4_-_83351005 | 3.85 |
ENST00000295470.5
|
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr17_+_16318850 | 3.85 |
ENST00000338560.7
|
TRPV2
|
transient receptor potential cation channel, subfamily V, member 2 |
| chr8_+_117778736 | 3.83 |
ENST00000309822.2
ENST00000357148.3 ENST00000517814.1 ENST00000517820.1 |
UTP23
|
UTP23, small subunit (SSU) processome component, homolog (yeast) |
| chr14_-_106994333 | 3.69 |
ENST00000390624.2
|
IGHV3-48
|
immunoglobulin heavy variable 3-48 |
| chr19_+_23299777 | 3.61 |
ENST00000597761.2
|
ZNF730
|
zinc finger protein 730 |
| chr19_-_12405689 | 3.59 |
ENST00000355684.5
|
ZNF44
|
zinc finger protein 44 |
| chr11_+_17298297 | 3.58 |
ENST00000529010.1
|
NUCB2
|
nucleobindin 2 |
| chr6_-_31697977 | 3.56 |
ENST00000375787.2
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr19_-_12405606 | 3.55 |
ENST00000356109.5
|
ZNF44
|
zinc finger protein 44 |
| chr4_-_174255536 | 3.53 |
ENST00000446922.2
|
HMGB2
|
high mobility group box 2 |
| chr5_+_162864575 | 3.52 |
ENST00000512163.1
ENST00000393929.1 ENST00000340828.2 ENST00000511683.2 ENST00000510097.1 ENST00000511490.2 ENST00000510664.1 |
CCNG1
|
cyclin G1 |
| chr19_-_6481776 | 3.47 |
ENST00000543576.1
ENST00000590173.1 ENST00000381480.2 |
DENND1C
|
DENN/MADD domain containing 1C |
| chr21_-_16437126 | 3.41 |
ENST00000318948.4
|
NRIP1
|
nuclear receptor interacting protein 1 |
| chr11_+_17298255 | 3.40 |
ENST00000531172.1
ENST00000533738.2 ENST00000323688.6 |
NUCB2
|
nucleobindin 2 |
| chr1_-_89458287 | 3.36 |
ENST00000370485.2
|
CCBL2
|
cysteine conjugate-beta lyase 2 |
| chr10_+_63661053 | 3.29 |
ENST00000279873.7
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like) |
| chr3_+_63953415 | 3.29 |
ENST00000484332.1
|
ATXN7
|
ataxin 7 |
| chr11_+_46403303 | 3.26 |
ENST00000407067.1
ENST00000395565.1 |
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr1_-_89458415 | 3.24 |
ENST00000321792.5
ENST00000370491.3 |
RBMXL1
CCBL2
|
RNA binding motif protein, X-linked-like 1 cysteine conjugate-beta lyase 2 |
| chr1_-_98386543 | 3.22 |
ENST00000423006.2
ENST00000370192.3 ENST00000306031.5 |
DPYD
|
dihydropyrimidine dehydrogenase |
| chr19_+_12721725 | 3.15 |
ENST00000446165.1
ENST00000343325.4 ENST00000458122.3 |
ZNF791
|
zinc finger protein 791 |
| chr9_+_131451480 | 3.12 |
ENST00000322030.8
|
SET
|
SET nuclear oncogene |
| chr18_+_21529811 | 2.97 |
ENST00000588004.1
|
LAMA3
|
laminin, alpha 3 |
| chr1_-_89458604 | 2.95 |
ENST00000260508.4
|
CCBL2
|
cysteine conjugate-beta lyase 2 |
| chr16_+_31366455 | 2.89 |
ENST00000268296.4
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr2_-_161349909 | 2.85 |
ENST00000392753.3
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1 |
| chr1_+_174843548 | 2.82 |
ENST00000478442.1
ENST00000465412.1 |
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr12_-_53045948 | 2.61 |
ENST00000309680.3
|
KRT2
|
keratin 2 |
| chr14_-_69445793 | 2.60 |
ENST00000538545.2
ENST00000394419.4 |
ACTN1
|
actinin, alpha 1 |
| chr11_+_46403194 | 2.58 |
ENST00000395569.4
ENST00000395566.4 |
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr17_+_7792101 | 2.57 |
ENST00000358181.4
ENST00000330494.7 |
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chr16_+_31366536 | 2.56 |
ENST00000562522.1
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr7_-_64023441 | 2.54 |
ENST00000309683.6
|
ZNF680
|
zinc finger protein 680 |
| chr6_+_84569359 | 2.51 |
ENST00000369681.5
ENST00000369679.4 |
CYB5R4
|
cytochrome b5 reductase 4 |
| chr22_-_36925186 | 2.49 |
ENST00000541106.1
ENST00000455547.1 ENST00000432675.1 |
EIF3D
|
eukaryotic translation initiation factor 3, subunit D |
| chr21_-_16437255 | 2.49 |
ENST00000400199.1
ENST00000400202.1 |
NRIP1
|
nuclear receptor interacting protein 1 |
| chr2_+_71295717 | 2.43 |
ENST00000418807.3
ENST00000443872.2 |
NAGK
|
N-acetylglucosamine kinase |
| chr14_+_64971292 | 2.42 |
ENST00000358738.3
ENST00000394712.2 |
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr1_+_113161778 | 2.37 |
ENST00000263168.3
|
CAPZA1
|
capping protein (actin filament) muscle Z-line, alpha 1 |
| chr22_-_39151463 | 2.36 |
ENST00000405510.1
ENST00000433561.1 |
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr22_-_36924944 | 2.30 |
ENST00000405442.1
ENST00000402116.1 |
EIF3D
|
eukaryotic translation initiation factor 3, subunit D |
| chr22_-_39151947 | 2.25 |
ENST00000216064.4
|
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr2_+_71295733 | 2.23 |
ENST00000443938.2
ENST00000244204.6 |
NAGK
|
N-acetylglucosamine kinase |
| chr20_+_44098346 | 2.20 |
ENST00000372676.3
|
WFDC2
|
WAP four-disulfide core domain 2 |
| chr3_+_184080387 | 2.19 |
ENST00000455712.1
|
POLR2H
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr12_-_66563831 | 2.18 |
ENST00000358230.3
|
TMBIM4
|
transmembrane BAX inhibitor motif containing 4 |
| chr20_+_44098385 | 2.17 |
ENST00000217425.5
ENST00000339946.3 |
WFDC2
|
WAP four-disulfide core domain 2 |
| chr6_-_32731243 | 2.15 |
ENST00000427449.1
ENST00000411527.1 |
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2 |
| chr11_-_62380199 | 2.07 |
ENST00000419857.1
ENST00000394773.2 |
EML3
|
echinoderm microtubule associated protein like 3 |
| chr1_+_9599540 | 2.05 |
ENST00000302692.6
|
SLC25A33
|
solute carrier family 25 (pyrimidine nucleotide carrier), member 33 |
| chr14_-_69445968 | 2.01 |
ENST00000438964.2
|
ACTN1
|
actinin, alpha 1 |
| chr14_+_93118813 | 1.93 |
ENST00000556418.1
|
RIN3
|
Ras and Rab interactor 3 |
| chr12_-_110883346 | 1.90 |
ENST00000547365.1
|
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr11_-_65374430 | 1.84 |
ENST00000532507.1
|
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11 |
| chr19_+_12175504 | 1.77 |
ENST00000439326.3
|
ZNF844
|
zinc finger protein 844 |
| chr8_+_55370487 | 1.77 |
ENST00000297316.4
|
SOX17
|
SRY (sex determining region Y)-box 17 |
| chr9_-_130829588 | 1.71 |
ENST00000373078.4
|
NAIF1
|
nuclear apoptosis inducing factor 1 |
| chr4_+_83351715 | 1.71 |
ENST00000273920.3
|
ENOPH1
|
enolase-phosphatase 1 |
| chr19_-_9929484 | 1.70 |
ENST00000586651.1
ENST00000586073.1 |
FBXL12
|
F-box and leucine-rich repeat protein 12 |
| chr11_+_65190245 | 1.69 |
ENST00000499732.1
ENST00000501122.2 ENST00000601801.1 |
NEAT1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
| chr4_-_76862117 | 1.69 |
ENST00000507956.1
ENST00000507187.2 ENST00000399497.3 ENST00000286733.4 |
NAAA
|
N-acylethanolamine acid amidase |
| chr14_-_69446034 | 1.64 |
ENST00000193403.6
|
ACTN1
|
actinin, alpha 1 |
| chr14_-_106471723 | 1.63 |
ENST00000390595.2
|
IGHV1-3
|
immunoglobulin heavy variable 1-3 |
| chr11_+_46366918 | 1.60 |
ENST00000528615.1
ENST00000395574.3 |
DGKZ
|
diacylglycerol kinase, zeta |
| chr20_+_62152077 | 1.56 |
ENST00000370179.3
ENST00000370177.1 |
PPDPF
|
pancreatic progenitor cell differentiation and proliferation factor |
| chr9_+_35829208 | 1.56 |
ENST00000439587.2
ENST00000377991.4 |
TMEM8B
|
transmembrane protein 8B |
| chr17_-_39191107 | 1.55 |
ENST00000344363.5
|
KRTAP1-3
|
keratin associated protein 1-3 |
| chr3_-_66551397 | 1.54 |
ENST00000383703.3
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr17_-_66287350 | 1.54 |
ENST00000580666.1
ENST00000583477.1 |
SLC16A6
|
solute carrier family 16, member 6 |
| chr1_-_207206092 | 1.49 |
ENST00000359470.5
ENST00000461135.2 |
C1orf116
|
chromosome 1 open reading frame 116 |
| chr1_-_155826967 | 1.48 |
ENST00000368331.1
ENST00000361040.5 ENST00000271883.5 |
GON4L
|
gon-4-like (C. elegans) |
| chr7_+_64363625 | 1.48 |
ENST00000476120.1
ENST00000319636.5 ENST00000545510.1 |
ZNF273
|
zinc finger protein 273 |
| chr17_-_74023474 | 1.47 |
ENST00000301607.3
|
EVPL
|
envoplakin |
| chr12_+_124118366 | 1.46 |
ENST00000539994.1
ENST00000538845.1 ENST00000228955.7 ENST00000543341.2 ENST00000536375.1 |
GTF2H3
|
general transcription factor IIH, polypeptide 3, 34kDa |
| chr19_+_19779619 | 1.45 |
ENST00000444249.2
ENST00000592502.1 |
ZNF101
|
zinc finger protein 101 |
| chr11_+_46402744 | 1.44 |
ENST00000533952.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr4_-_2264015 | 1.40 |
ENST00000337190.2
|
MXD4
|
MAX dimerization protein 4 |
| chr17_-_39928106 | 1.38 |
ENST00000540235.1
|
JUP
|
junction plakoglobin |
| chr12_-_124118296 | 1.35 |
ENST00000424014.2
ENST00000537073.1 |
EIF2B1
|
eukaryotic translation initiation factor 2B, subunit 1 alpha, 26kDa |
| chr19_-_9929708 | 1.35 |
ENST00000247977.4
ENST00000590277.1 ENST00000588922.1 ENST00000589626.1 ENST00000592067.1 ENST00000586469.1 |
FBXL12
|
F-box and leucine-rich repeat protein 12 |
| chr1_-_226129189 | 1.31 |
ENST00000366820.5
|
LEFTY2
|
left-right determination factor 2 |
| chrX_+_49687267 | 1.28 |
ENST00000376091.3
|
CLCN5
|
chloride channel, voltage-sensitive 5 |
| chr12_+_56473910 | 1.26 |
ENST00000411731.2
|
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr5_+_177631523 | 1.22 |
ENST00000506339.1
ENST00000355836.5 ENST00000514633.1 ENST00000515193.1 ENST00000506259.1 ENST00000504898.1 |
HNRNPAB
|
heterogeneous nuclear ribonucleoprotein A/B |
| chr3_-_66551351 | 1.21 |
ENST00000273261.3
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr10_-_103880209 | 1.20 |
ENST00000425280.1
|
LDB1
|
LIM domain binding 1 |
| chr3_+_184081213 | 1.19 |
ENST00000429568.1
|
POLR2H
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr8_-_103876965 | 1.18 |
ENST00000337198.5
|
AZIN1
|
antizyme inhibitor 1 |
| chr3_+_184080790 | 1.18 |
ENST00000430783.1
|
POLR2H
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr2_+_11886710 | 1.17 |
ENST00000256720.2
ENST00000441684.1 ENST00000423495.1 |
LPIN1
|
lipin 1 |
| chr5_+_177631497 | 1.16 |
ENST00000358344.3
|
HNRNPAB
|
heterogeneous nuclear ribonucleoprotein A/B |
| chr7_+_77167376 | 1.13 |
ENST00000435495.2
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr22_+_40440804 | 1.12 |
ENST00000441751.1
ENST00000301923.9 |
TNRC6B
|
trinucleotide repeat containing 6B |
| chr8_-_144897549 | 1.10 |
ENST00000356994.2
ENST00000320476.3 |
SCRIB
|
scribbled planar cell polarity protein |
| chr12_+_56473939 | 1.08 |
ENST00000450146.2
|
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr1_-_226129083 | 1.04 |
ENST00000420304.2
|
LEFTY2
|
left-right determination factor 2 |
| chr17_-_66287257 | 1.03 |
ENST00000327268.4
|
SLC16A6
|
solute carrier family 16, member 6 |
| chrX_-_154688276 | 1.02 |
ENST00000369445.2
|
F8A3
|
coagulation factor VIII-associated 3 |
| chr9_+_132815985 | 1.01 |
ENST00000372410.3
|
GPR107
|
G protein-coupled receptor 107 |
| chr3_+_184081175 | 1.00 |
ENST00000452961.1
ENST00000296223.3 |
POLR2H
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr17_-_60142609 | 0.98 |
ENST00000397786.2
|
MED13
|
mediator complex subunit 13 |
| chr2_-_10588630 | 0.97 |
ENST00000234111.4
|
ODC1
|
ornithine decarboxylase 1 |
| chr8_-_144897138 | 0.96 |
ENST00000377533.3
|
SCRIB
|
scribbled planar cell polarity protein |
| chr15_+_41136216 | 0.93 |
ENST00000562057.1
ENST00000344051.4 |
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr9_-_127269661 | 0.93 |
ENST00000373588.4
|
NR5A1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr11_-_62323702 | 0.92 |
ENST00000530285.1
|
AHNAK
|
AHNAK nucleoprotein |
| chr1_+_151512775 | 0.87 |
ENST00000368849.3
ENST00000392712.3 ENST00000353024.3 ENST00000368848.2 ENST00000538902.1 |
TUFT1
|
tuftelin 1 |
| chr17_-_42452063 | 0.84 |
ENST00000588098.1
|
ITGA2B
|
integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) |
| chr12_-_4554780 | 0.78 |
ENST00000228837.2
|
FGF6
|
fibroblast growth factor 6 |
| chr22_+_31489344 | 0.77 |
ENST00000404574.1
|
SMTN
|
smoothelin |
| chr17_-_1532106 | 0.75 |
ENST00000301335.5
ENST00000382147.4 |
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr3_+_120461484 | 0.75 |
ENST00000484715.1
ENST00000469772.1 ENST00000283875.5 ENST00000492959.1 |
GTF2E1
|
general transcription factor IIE, polypeptide 1, alpha 56kDa |
| chr19_+_44100727 | 0.74 |
ENST00000528387.1
ENST00000529930.1 ENST00000336564.4 ENST00000607544.1 ENST00000526798.1 |
ZNF576
SRRM5
|
zinc finger protein 576 serine/arginine repetitive matrix 5 |
| chr3_-_48672859 | 0.74 |
ENST00000395550.2
ENST00000455886.2 ENST00000431739.1 ENST00000426599.1 ENST00000383733.3 ENST00000420764.2 ENST00000337000.8 |
SLC26A6
|
solute carrier family 26 (anion exchanger), member 6 |
| chr19_+_48969094 | 0.73 |
ENST00000595676.1
|
CTC-273B12.7
|
Uncharacterized protein |
| chr19_+_44100632 | 0.72 |
ENST00000533118.1
|
ZNF576
|
zinc finger protein 576 |
| chr9_-_129884902 | 0.69 |
ENST00000373417.1
|
ANGPTL2
|
angiopoietin-like 2 |
| chr20_+_61867235 | 0.69 |
ENST00000342412.6
ENST00000217169.3 |
BIRC7
|
baculoviral IAP repeat containing 7 |
| chr4_+_47487285 | 0.66 |
ENST00000273859.3
ENST00000504445.1 |
ATP10D
|
ATPase, class V, type 10D |
| chr1_-_28241226 | 0.64 |
ENST00000373912.3
ENST00000373909.3 |
RPA2
|
replication protein A2, 32kDa |
| chr1_-_155959853 | 0.63 |
ENST00000462460.2
ENST00000368316.1 |
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr19_+_12075844 | 0.63 |
ENST00000592625.1
ENST00000586494.1 ENST00000343949.5 ENST00000545530.1 ENST00000358987.3 |
ZNF763
|
zinc finger protein 763 |
| chr2_+_241544834 | 0.63 |
ENST00000319838.5
ENST00000403859.1 ENST00000438013.2 |
GPR35
|
G protein-coupled receptor 35 |
| chr3_+_184081137 | 0.62 |
ENST00000443489.1
|
POLR2H
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr11_-_123612319 | 0.61 |
ENST00000526252.1
ENST00000530393.1 ENST00000533463.1 ENST00000336139.4 ENST00000529691.1 ENST00000528306.1 |
ZNF202
|
zinc finger protein 202 |
| chr5_-_148758839 | 0.61 |
ENST00000261796.3
|
IL17B
|
interleukin 17B |
| chr3_-_50605077 | 0.60 |
ENST00000426034.1
ENST00000441239.1 |
C3orf18
|
chromosome 3 open reading frame 18 |
| chr2_+_241564655 | 0.59 |
ENST00000407714.1
|
GPR35
|
G protein-coupled receptor 35 |
| chr20_+_44509857 | 0.59 |
ENST00000372523.1
ENST00000372520.1 |
ZSWIM1
|
zinc finger, SWIM-type containing 1 |
| chr3_+_40547483 | 0.58 |
ENST00000420891.1
ENST00000314529.6 ENST00000418905.1 |
ZNF620
|
zinc finger protein 620 |
| chr12_-_66563786 | 0.57 |
ENST00000542724.1
|
TMBIM4
|
transmembrane BAX inhibitor motif containing 4 |
| chr17_+_57642886 | 0.57 |
ENST00000251241.4
ENST00000451169.2 ENST00000425628.3 ENST00000584385.1 ENST00000580030.1 |
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr7_+_43966018 | 0.55 |
ENST00000222402.3
ENST00000446008.1 ENST00000394798.4 |
UBE2D4
|
ubiquitin-conjugating enzyme E2D 4 (putative) |
| chr11_+_31531291 | 0.55 |
ENST00000350638.5
ENST00000379163.5 ENST00000395934.2 |
ELP4
|
elongator acetyltransferase complex subunit 4 |
| chr9_-_129885010 | 0.53 |
ENST00000373425.3
|
ANGPTL2
|
angiopoietin-like 2 |
| chr7_+_136553824 | 0.53 |
ENST00000320658.5
ENST00000453373.1 ENST00000397608.3 ENST00000402486.3 ENST00000401861.1 |
CHRM2
|
cholinergic receptor, muscarinic 2 |
| chr2_+_74056066 | 0.53 |
ENST00000339566.3
ENST00000409707.1 ENST00000452725.1 ENST00000432295.2 ENST00000424659.1 ENST00000394073.1 |
STAMBP
|
STAM binding protein |
| chr5_-_148929848 | 0.53 |
ENST00000504676.1
ENST00000515435.1 |
CSNK1A1
|
casein kinase 1, alpha 1 |
| chr2_+_130737223 | 0.51 |
ENST00000410061.2
|
RAB6C
|
RAB6C, member RAS oncogene family |
| chr2_-_219157250 | 0.50 |
ENST00000434015.2
ENST00000444183.1 ENST00000420341.1 ENST00000453281.1 ENST00000258412.3 ENST00000440422.1 |
TMBIM1
|
transmembrane BAX inhibitor motif containing 1 |
| chr7_+_136553370 | 0.46 |
ENST00000445907.2
|
CHRM2
|
cholinergic receptor, muscarinic 2 |
| chr6_-_31080336 | 0.45 |
ENST00000259870.3
|
C6orf15
|
chromosome 6 open reading frame 15 |
| chr12_+_41582103 | 0.45 |
ENST00000402685.2
|
PDZRN4
|
PDZ domain containing ring finger 4 |
| chr11_+_67776012 | 0.45 |
ENST00000539229.1
|
ALDH3B1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr21_-_28338732 | 0.45 |
ENST00000284987.5
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
| chr2_+_74056147 | 0.44 |
ENST00000394070.2
ENST00000536064.1 |
STAMBP
|
STAM binding protein |
| chr12_-_121712313 | 0.43 |
ENST00000392474.2
|
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta |
| chr19_-_6375860 | 0.42 |
ENST00000245810.1
|
PSPN
|
persephin |
| chr15_-_90233866 | 0.41 |
ENST00000561257.1
|
PEX11A
|
peroxisomal biogenesis factor 11 alpha |
| chr9_+_91606355 | 0.40 |
ENST00000358157.2
|
S1PR3
|
sphingosine-1-phosphate receptor 3 |
| chr14_-_61124977 | 0.39 |
ENST00000554986.1
|
SIX1
|
SIX homeobox 1 |
| chrX_+_69674943 | 0.39 |
ENST00000542398.1
|
DLG3
|
discs, large homolog 3 (Drosophila) |
| chr4_-_57301748 | 0.36 |
ENST00000264220.2
|
PPAT
|
phosphoribosyl pyrophosphate amidotransferase |
| chr1_+_186265399 | 0.33 |
ENST00000367486.3
ENST00000367484.3 ENST00000533951.1 ENST00000367482.4 ENST00000367483.4 ENST00000367485.4 ENST00000445192.2 |
PRG4
|
proteoglycan 4 |
| chr2_+_233415363 | 0.33 |
ENST00000409514.1
ENST00000409098.1 ENST00000409495.1 ENST00000409167.3 ENST00000409322.1 ENST00000409394.1 |
EIF4E2
|
eukaryotic translation initiation factor 4E family member 2 |
| chr11_-_77122928 | 0.32 |
ENST00000528203.1
ENST00000528592.1 ENST00000528633.1 ENST00000529248.1 |
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr10_+_99344104 | 0.30 |
ENST00000555577.1
ENST00000370649.3 |
PI4K2A
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha Phosphatidylinositol 4-kinase type 2-alpha; Uncharacterized protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of TFCP2L1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 18.1 | GO:0046102 | hypoxanthine salvage(GO:0043103) inosine metabolic process(GO:0046102) |
| 2.4 | 18.9 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 2.3 | 9.2 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 2.1 | 6.2 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 2.0 | 13.8 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 1.6 | 4.8 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 1.2 | 6.2 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 1.2 | 7.3 | GO:0030421 | defecation(GO:0030421) |
| 1.2 | 4.7 | GO:0006051 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 1.1 | 3.2 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) beta-alanine metabolic process(GO:0019482) |
| 0.9 | 2.7 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.9 | 6.3 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.8 | 5.0 | GO:0043366 | beta selection(GO:0043366) |
| 0.8 | 6.3 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.8 | 4.6 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.7 | 2.1 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.7 | 2.0 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.6 | 3.8 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.6 | 1.8 | GO:0009996 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) negative regulation of cell fate specification(GO:0009996) gall bladder development(GO:0061010) |
| 0.6 | 13.6 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.5 | 2.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.4 | 8.9 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.4 | 5.9 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.4 | 6.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.4 | 6.4 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.4 | 3.5 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.3 | 3.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.3 | 1.4 | GO:0002159 | desmosome assembly(GO:0002159) endothelial cell-cell adhesion(GO:0071603) |
| 0.3 | 1.8 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.3 | 0.8 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.3 | 3.6 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.2 | 0.7 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.2 | 0.9 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.2 | 1.6 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.2 | 15.1 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.2 | 4.6 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.2 | 1.2 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.2 | 1.2 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.2 | 1.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.2 | 1.7 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.2 | 0.6 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.2 | 1.0 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.2 | 2.6 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.1 | 25.5 | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.1 | 0.4 | GO:0061055 | myotome development(GO:0061055) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 0.5 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.1 | 10.2 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.1 | 8.0 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 12.2 | GO:0032945 | negative regulation of mononuclear cell proliferation(GO:0032945) negative regulation of lymphocyte proliferation(GO:0050672) |
| 0.1 | 3.1 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 6.2 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.1 | 0.4 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.1 | 3.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.1 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.1 | 0.3 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.1 | 0.1 | GO:0072179 | nephric duct formation(GO:0072179) |
| 0.1 | 3.3 | GO:0016577 | histone demethylation(GO:0016577) |
| 0.1 | 1.0 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.1 | 6.7 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.1 | 1.1 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 0.4 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 0.5 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.1 | 0.9 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 0.4 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 1.1 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 11.7 | GO:0043542 | endothelial cell migration(GO:0043542) |
| 0.1 | 5.2 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.4 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.6 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.4 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 4.0 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.7 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 2.5 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 2.8 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 1.5 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 1.6 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 1.4 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 1.3 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.6 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 2.6 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 1.0 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.7 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 1.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.9 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 1.7 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 2.4 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 11.7 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.2 | GO:2000675 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) determination of digestive tract left/right asymmetry(GO:0071907) determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 0.9 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 1.5 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 8.7 | GO:0043312 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.4 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.0 | 0.5 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 1.6 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.9 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 5.7 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 1.8 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.3 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.2 | GO:0034776 | response to histamine(GO:0034776) cellular response to histamine(GO:0071420) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 8.8 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.9 | 6.5 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.7 | 2.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.5 | 7.6 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.5 | 15.7 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.5 | 1.5 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.4 | 8.9 | GO:0031105 | septin complex(GO:0031105) |
| 0.4 | 3.0 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.4 | 4.6 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.3 | 6.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.3 | 2.4 | GO:0071203 | WASH complex(GO:0071203) |
| 0.3 | 6.7 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 1.0 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.2 | 1.4 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.2 | 5.0 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 14.0 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.1 | 3.8 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 6.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.9 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 15.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.3 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 2.6 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 3.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.3 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.1 | 0.7 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 4.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 0.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 13.8 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 16.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 6.4 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.7 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.6 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 3.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 13.5 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 4.5 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 1.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 4.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 3.4 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 5.5 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.9 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.3 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 17.3 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 1.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.9 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 1.9 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 12.1 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 3.2 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.7 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 1.4 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.2 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 3.1 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 9.1 | GO:0030529 | intracellular ribonucleoprotein complex(GO:0030529) |
| 0.0 | 3.9 | GO:0005874 | microtubule(GO:0005874) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 18.1 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 2.1 | 6.2 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 1.8 | 31.6 | GO:0019864 | IgG binding(GO:0019864) |
| 1.6 | 6.3 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 1.6 | 4.7 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 1.4 | 15.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 1.1 | 4.4 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.9 | 3.5 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.8 | 13.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.7 | 3.6 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.7 | 6.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.7 | 2.0 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.5 | 3.8 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.5 | 6.7 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.5 | 1.8 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.4 | 8.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.4 | 12.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.4 | 1.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.3 | 6.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.3 | 1.0 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.3 | 4.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.2 | 6.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.2 | 12.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 5.9 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.2 | 4.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 5.0 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.2 | 0.4 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.2 | 0.8 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.2 | 2.5 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.2 | 1.0 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.2 | 0.7 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.2 | 1.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.2 | 7.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 1.4 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 1.7 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.9 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.1 | 1.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 3.3 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 3.2 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 1.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 5.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 1.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.3 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.1 | 1.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 3.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 14.6 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 2.5 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 7.9 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 1.0 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 0.6 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 2.6 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 0.5 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 5.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.9 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 4.9 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 2.6 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.4 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 7.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 3.5 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.8 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.5 | GO:0008135 | translation factor activity, RNA binding(GO:0008135) |
| 0.0 | 0.4 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 2.9 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 1.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.3 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.2 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 1.8 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.5 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 6.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.8 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 31.6 | GO:0003677 | DNA binding(GO:0003677) |
| 0.0 | 0.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.4 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 17.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.5 | 31.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.3 | 30.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 10.6 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.2 | 6.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 5.2 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 5.9 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 6.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 7.2 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 5.0 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 3.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 1.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 4.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 4.1 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.4 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 1.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 2.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 11.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 2.6 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 2.5 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.8 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 13.8 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 1.0 | 13.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.7 | 18.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.3 | 6.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.2 | 3.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 5.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.2 | 6.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 8.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.2 | 17.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.2 | 7.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.2 | 3.5 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 7.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 6.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 8.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 2.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 14.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 2.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 0.6 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 1.1 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.1 | 0.8 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 4.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.8 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 1.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 4.6 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 2.4 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 1.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.5 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 2.7 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 1.2 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 1.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |