Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
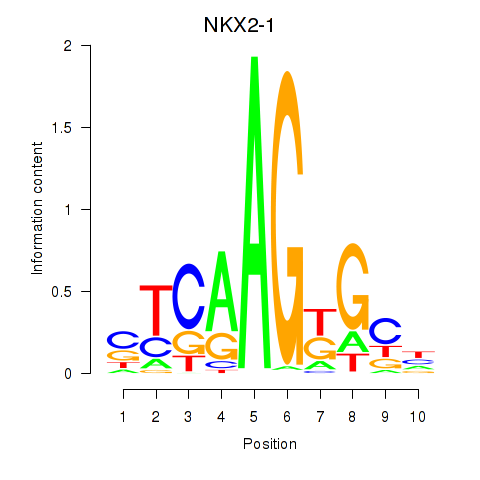
Results for NKX2-1
Z-value: 0.06
Transcription factors associated with NKX2-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-1
|
ENSG00000136352.13 | NK2 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-1 | hg19_v2_chr14_-_36989336_36989354 | 0.05 | 4.6e-01 | Click! |
Activity profile of NKX2-1 motif
Sorted Z-values of NKX2-1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_60223225 | 12.50 |
ENST00000524807.1
ENST00000345732.4 |
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr11_+_60223312 | 12.31 |
ENST00000532491.1
ENST00000532073.1 ENST00000534668.1 ENST00000528313.1 ENST00000533306.1 |
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr19_+_1065922 | 11.50 |
ENST00000539243.2
|
HMHA1
|
histocompatibility (minor) HA-1 |
| chr13_-_41240717 | 8.24 |
ENST00000379561.5
|
FOXO1
|
forkhead box O1 |
| chr1_-_153517473 | 8.11 |
ENST00000368715.1
|
S100A4
|
S100 calcium binding protein A4 |
| chr15_+_89182178 | 7.64 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr12_+_25205568 | 7.49 |
ENST00000548766.1
ENST00000556887.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr12_+_25205666 | 7.45 |
ENST00000547044.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr1_-_153518270 | 6.91 |
ENST00000354332.4
ENST00000368716.4 |
S100A4
|
S100 calcium binding protein A4 |
| chr12_+_25205446 | 6.77 |
ENST00000557489.1
ENST00000354454.3 ENST00000536173.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr15_+_89181974 | 6.72 |
ENST00000306072.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chrX_+_131157290 | 6.38 |
ENST00000394334.2
|
MST4
|
Serine/threonine-protein kinase MST4 |
| chrX_+_131157322 | 6.37 |
ENST00000481105.1
ENST00000354719.6 ENST00000394335.2 |
MST4
|
Serine/threonine-protein kinase MST4 |
| chr1_-_25291475 | 6.24 |
ENST00000338888.3
ENST00000399916.1 |
RUNX3
|
runt-related transcription factor 3 |
| chr5_-_171615315 | 6.19 |
ENST00000176763.5
|
STK10
|
serine/threonine kinase 10 |
| chr15_+_89182156 | 5.51 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr10_-_98031310 | 4.73 |
ENST00000427367.2
ENST00000413476.2 |
BLNK
|
B-cell linker |
| chr3_-_16524357 | 4.57 |
ENST00000432519.1
|
RFTN1
|
raftlin, lipid raft linker 1 |
| chr17_+_4613918 | 4.32 |
ENST00000574954.1
ENST00000346341.2 ENST00000572457.1 ENST00000381488.6 ENST00000412477.3 ENST00000571428.1 ENST00000575877.1 |
ARRB2
|
arrestin, beta 2 |
| chr10_-_98031265 | 4.04 |
ENST00000224337.5
ENST00000371176.2 |
BLNK
|
B-cell linker |
| chr19_+_24009879 | 3.90 |
ENST00000354585.4
|
RPSAP58
|
ribosomal protein SA pseudogene 58 |
| chr11_-_104827425 | 3.80 |
ENST00000393150.3
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr17_-_76778339 | 3.78 |
ENST00000591455.1
ENST00000446868.3 ENST00000361101.4 ENST00000589296.1 |
CYTH1
|
cytohesin 1 |
| chr12_-_21810726 | 3.67 |
ENST00000396076.1
|
LDHB
|
lactate dehydrogenase B |
| chr12_-_21810765 | 3.66 |
ENST00000450584.1
ENST00000350669.1 |
LDHB
|
lactate dehydrogenase B |
| chr1_+_24019099 | 3.49 |
ENST00000443624.1
ENST00000458455.1 |
RPL11
|
ribosomal protein L11 |
| chr17_+_46048376 | 3.47 |
ENST00000338399.4
|
CDK5RAP3
|
CDK5 regulatory subunit associated protein 3 |
| chr19_-_36231437 | 3.26 |
ENST00000591748.1
|
IGFLR1
|
IGF-like family receptor 1 |
| chr6_+_31554779 | 3.01 |
ENST00000376090.2
|
LST1
|
leukocyte specific transcript 1 |
| chr16_-_67190152 | 2.88 |
ENST00000486556.1
|
TRADD
|
TNFRSF1A-associated via death domain |
| chr1_-_38397384 | 2.67 |
ENST00000373027.1
|
INPP5B
|
inositol polyphosphate-5-phosphatase, 75kDa |
| chr15_-_44955842 | 2.67 |
ENST00000427534.2
ENST00000559193.1 ENST00000261866.7 ENST00000535302.2 ENST00000558319.1 |
SPG11
|
spastic paraplegia 11 (autosomal recessive) |
| chr8_-_16859690 | 2.62 |
ENST00000180166.5
|
FGF20
|
fibroblast growth factor 20 |
| chr1_+_15736359 | 2.54 |
ENST00000375980.4
|
EFHD2
|
EF-hand domain family, member D2 |
| chr6_-_26056695 | 2.30 |
ENST00000343677.2
|
HIST1H1C
|
histone cluster 1, H1c |
| chr19_-_46234119 | 2.30 |
ENST00000317683.3
|
FBXO46
|
F-box protein 46 |
| chr5_+_134094461 | 2.25 |
ENST00000452510.2
ENST00000354283.4 |
DDX46
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 |
| chr16_+_1359138 | 2.21 |
ENST00000325437.5
|
UBE2I
|
ubiquitin-conjugating enzyme E2I |
| chr11_+_35160709 | 2.20 |
ENST00000415148.2
ENST00000433354.2 ENST00000449691.2 ENST00000437706.2 ENST00000360158.4 ENST00000428726.2 ENST00000526669.2 ENST00000433892.2 ENST00000278386.6 ENST00000434472.2 ENST00000352818.4 ENST00000442151.2 |
CD44
|
CD44 molecule (Indian blood group) |
| chrX_-_70474499 | 2.20 |
ENST00000353904.2
|
ZMYM3
|
zinc finger, MYM-type 3 |
| chr8_-_144679296 | 2.18 |
ENST00000317198.6
|
EEF1D
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr6_-_137113604 | 2.18 |
ENST00000359015.4
|
MAP3K5
|
mitogen-activated protein kinase kinase kinase 5 |
| chr12_+_121837905 | 2.03 |
ENST00000392465.3
ENST00000554606.1 ENST00000392464.2 ENST00000555076.1 |
RNF34
|
ring finger protein 34, E3 ubiquitin protein ligase |
| chr14_-_58893832 | 1.99 |
ENST00000556007.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr15_+_40453204 | 1.99 |
ENST00000287598.6
ENST00000412359.3 |
BUB1B
|
BUB1 mitotic checkpoint serine/threonine kinase B |
| chr12_-_54694758 | 1.98 |
ENST00000553070.1
|
NFE2
|
nuclear factor, erythroid 2 |
| chr16_-_28222797 | 1.92 |
ENST00000569951.1
ENST00000565698.1 |
XPO6
|
exportin 6 |
| chr8_-_28747424 | 1.91 |
ENST00000523436.1
ENST00000397363.4 ENST00000521777.1 ENST00000520184.1 ENST00000521022.1 |
INTS9
|
integrator complex subunit 9 |
| chr3_-_52029830 | 1.87 |
ENST00000492277.1
ENST00000475248.1 ENST00000479017.1 ENST00000495383.1 ENST00000466397.1 ENST00000481629.1 |
RPL29
|
ribosomal protein L29 |
| chr15_-_44116873 | 1.74 |
ENST00000267812.3
|
MFAP1
|
microfibrillar-associated protein 1 |
| chr16_+_71929397 | 1.66 |
ENST00000537613.1
ENST00000424485.2 ENST00000606369.1 ENST00000329908.8 ENST00000538850.1 ENST00000541918.1 ENST00000534994.1 ENST00000378798.5 ENST00000539186.1 |
IST1
|
increased sodium tolerance 1 homolog (yeast) |
| chr19_-_10679644 | 1.53 |
ENST00000393599.2
|
CDKN2D
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr3_-_52029958 | 1.50 |
ENST00000294189.6
|
RPL29
|
ribosomal protein L29 |
| chr1_-_92952433 | 1.49 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr18_-_24283586 | 1.48 |
ENST00000579458.1
|
U3
|
Small nucleolar RNA U3 |
| chr3_+_4535025 | 1.48 |
ENST00000302640.8
ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr11_+_105948216 | 1.45 |
ENST00000278618.4
|
AASDHPPT
|
aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase |
| chr16_-_58663720 | 1.43 |
ENST00000564557.1
ENST00000569240.1 ENST00000441024.2 ENST00000569020.1 ENST00000317147.5 |
CNOT1
|
CCR4-NOT transcription complex, subunit 1 |
| chr2_+_99797542 | 1.43 |
ENST00000338148.3
ENST00000512183.2 |
MRPL30
C2orf15
|
mitochondrial ribosomal protein L30 chromosome 2 open reading frame 15 |
| chr12_-_719573 | 1.36 |
ENST00000397265.3
|
NINJ2
|
ninjurin 2 |
| chr3_+_23847432 | 1.36 |
ENST00000346855.3
|
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr11_-_62313090 | 1.35 |
ENST00000528508.1
ENST00000533365.1 |
AHNAK
|
AHNAK nucleoprotein |
| chr1_-_175712665 | 1.35 |
ENST00000263525.2
|
TNR
|
tenascin R |
| chrX_-_70473957 | 1.34 |
ENST00000373984.3
ENST00000314425.5 ENST00000373982.1 |
ZMYM3
|
zinc finger, MYM-type 3 |
| chr10_+_111967345 | 1.31 |
ENST00000332674.5
ENST00000453116.1 |
MXI1
|
MAX interactor 1, dimerization protein |
| chr15_+_57511609 | 1.30 |
ENST00000543579.1
ENST00000537840.1 ENST00000343827.3 |
TCF12
|
transcription factor 12 |
| chr4_+_144354644 | 1.30 |
ENST00000512843.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr22_+_24891210 | 1.28 |
ENST00000382760.2
|
UPB1
|
ureidopropionase, beta |
| chr6_-_55739542 | 1.23 |
ENST00000446683.2
|
BMP5
|
bone morphogenetic protein 5 |
| chr14_+_74353320 | 1.22 |
ENST00000540593.1
ENST00000555730.1 |
ZNF410
|
zinc finger protein 410 |
| chr17_-_42466864 | 1.17 |
ENST00000353281.4
ENST00000262407.5 |
ITGA2B
|
integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) |
| chr6_-_27841289 | 1.17 |
ENST00000355981.2
|
HIST1H4L
|
histone cluster 1, H4l |
| chr16_-_28223166 | 1.13 |
ENST00000304658.5
|
XPO6
|
exportin 6 |
| chr1_+_32930647 | 1.09 |
ENST00000609129.1
|
ZBTB8B
|
zinc finger and BTB domain containing 8B |
| chrX_-_110655306 | 1.08 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr15_+_77287426 | 1.07 |
ENST00000558012.1
ENST00000267939.5 ENST00000379595.3 |
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1 |
| chr1_+_206643787 | 0.90 |
ENST00000367120.3
|
IKBKE
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr21_+_45432174 | 0.89 |
ENST00000380221.3
ENST00000291574.4 |
TRAPPC10
|
trafficking protein particle complex 10 |
| chr19_+_48958766 | 0.87 |
ENST00000342291.2
|
KCNJ14
|
potassium inwardly-rectifying channel, subfamily J, member 14 |
| chr12_+_41582103 | 0.79 |
ENST00000402685.2
|
PDZRN4
|
PDZ domain containing ring finger 4 |
| chr18_+_42260861 | 0.73 |
ENST00000282030.5
|
SETBP1
|
SET binding protein 1 |
| chr12_+_60058458 | 0.73 |
ENST00000548610.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr11_-_65325430 | 0.69 |
ENST00000322147.4
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr6_+_42883727 | 0.69 |
ENST00000304672.1
ENST00000441198.1 ENST00000446507.1 |
PTCRA
|
pre T-cell antigen receptor alpha |
| chr22_-_30968813 | 0.68 |
ENST00000443111.2
ENST00000443136.1 ENST00000426220.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr3_-_11685345 | 0.65 |
ENST00000430365.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr17_-_47492164 | 0.62 |
ENST00000512041.2
ENST00000446735.1 ENST00000504124.1 |
PHB
|
prohibitin |
| chr1_-_160232197 | 0.60 |
ENST00000419626.1
ENST00000610139.1 ENST00000475733.1 ENST00000407642.2 ENST00000368073.3 ENST00000326837.2 |
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr15_-_66790146 | 0.59 |
ENST00000316634.5
|
SNAPC5
|
small nuclear RNA activating complex, polypeptide 5, 19kDa |
| chr1_-_153538011 | 0.59 |
ENST00000368707.4
|
S100A2
|
S100 calcium binding protein A2 |
| chr11_+_66512303 | 0.58 |
ENST00000532565.2
|
C11orf80
|
chromosome 11 open reading frame 80 |
| chr6_-_55740352 | 0.53 |
ENST00000370830.3
|
BMP5
|
bone morphogenetic protein 5 |
| chr8_+_94752349 | 0.53 |
ENST00000391680.1
|
RBM12B-AS1
|
RBM12B antisense RNA 1 |
| chr19_-_51336443 | 0.53 |
ENST00000598673.1
|
KLK15
|
kallikrein-related peptidase 15 |
| chr17_+_76210367 | 0.52 |
ENST00000592734.1
ENST00000587746.1 |
BIRC5
|
baculoviral IAP repeat containing 5 |
| chr11_+_66512089 | 0.51 |
ENST00000524551.1
ENST00000525908.1 ENST00000360962.4 ENST00000346672.4 ENST00000527634.1 ENST00000540737.1 |
C11orf80
|
chromosome 11 open reading frame 80 |
| chr10_+_102222798 | 0.50 |
ENST00000343737.5
|
WNT8B
|
wingless-type MMTV integration site family, member 8B |
| chr2_-_157189180 | 0.49 |
ENST00000539077.1
ENST00000424077.1 ENST00000426264.1 ENST00000339562.4 ENST00000421709.1 |
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chrX_-_32173579 | 0.48 |
ENST00000359836.1
ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD
|
dystrophin |
| chr2_+_136289030 | 0.46 |
ENST00000409478.1
ENST00000264160.4 ENST00000329971.3 ENST00000438014.1 |
R3HDM1
|
R3H domain containing 1 |
| chr2_+_119699742 | 0.45 |
ENST00000327097.4
|
MARCO
|
macrophage receptor with collagenous structure |
| chr17_-_47492236 | 0.43 |
ENST00000434917.2
ENST00000300408.3 ENST00000511832.1 ENST00000419140.2 |
PHB
|
prohibitin |
| chr20_-_22565101 | 0.42 |
ENST00000419308.2
|
FOXA2
|
forkhead box A2 |
| chrX_-_134478012 | 0.42 |
ENST00000370766.3
|
ZNF75D
|
zinc finger protein 75D |
| chr12_+_50478977 | 0.42 |
ENST00000381513.4
|
SMARCD1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr4_-_54930790 | 0.41 |
ENST00000263921.3
|
CHIC2
|
cysteine-rich hydrophobic domain 2 |
| chr9_-_128246769 | 0.41 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr2_-_31360887 | 0.40 |
ENST00000420311.2
ENST00000356174.3 ENST00000324589.5 |
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr8_+_22019168 | 0.40 |
ENST00000318561.3
ENST00000521315.1 ENST00000437090.2 ENST00000520605.1 ENST00000522109.1 ENST00000524255.1 ENST00000523296.1 ENST00000518615.1 |
SFTPC
|
surfactant protein C |
| chr22_-_30925150 | 0.39 |
ENST00000437871.1
|
SEC14L6
|
SEC14-like 6 (S. cerevisiae) |
| chr12_+_121416489 | 0.36 |
ENST00000541395.1
ENST00000544413.1 |
HNF1A
|
HNF1 homeobox A |
| chr18_-_58040000 | 0.36 |
ENST00000299766.3
|
MC4R
|
melanocortin 4 receptor |
| chrX_+_153170189 | 0.36 |
ENST00000358927.2
|
AVPR2
|
arginine vasopressin receptor 2 |
| chr1_-_153538292 | 0.33 |
ENST00000497140.1
ENST00000368708.3 |
S100A2
|
S100 calcium binding protein A2 |
| chr7_-_27224842 | 0.31 |
ENST00000517402.1
|
HOXA11
|
homeobox A11 |
| chr4_+_55524085 | 0.30 |
ENST00000412167.2
ENST00000288135.5 |
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
| chr7_-_27224795 | 0.28 |
ENST00000006015.3
|
HOXA11
|
homeobox A11 |
| chr14_+_81421921 | 0.27 |
ENST00000554263.1
ENST00000554435.1 |
TSHR
|
thyroid stimulating hormone receptor |
| chr19_+_55014085 | 0.27 |
ENST00000351841.2
|
LAIR2
|
leukocyte-associated immunoglobulin-like receptor 2 |
| chr1_+_55464600 | 0.27 |
ENST00000371265.4
|
BSND
|
Bartter syndrome, infantile, with sensorineural deafness (Barttin) |
| chr17_+_59529743 | 0.26 |
ENST00000589003.1
ENST00000393853.4 |
TBX4
|
T-box 4 |
| chr2_+_119699864 | 0.24 |
ENST00000541757.1
ENST00000412481.1 |
MARCO
|
macrophage receptor with collagenous structure |
| chr12_-_67197760 | 0.24 |
ENST00000539540.1
ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr1_+_206643806 | 0.19 |
ENST00000537984.1
|
IKBKE
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr10_-_81320151 | 0.19 |
ENST00000372325.2
ENST00000372327.5 ENST00000417041.1 |
SFTPA2
|
surfactant protein A2 |
| chrX_+_78003204 | 0.16 |
ENST00000435339.3
ENST00000514744.1 |
LPAR4
|
lysophosphatidic acid receptor 4 |
| chr17_+_76210267 | 0.13 |
ENST00000301633.4
ENST00000350051.3 ENST00000374948.2 ENST00000590449.1 |
BIRC5
|
baculoviral IAP repeat containing 5 |
| chr6_+_31674639 | 0.13 |
ENST00000556581.1
ENST00000375832.4 ENST00000503322.1 |
LY6G6F
MEGT1
|
lymphocyte antigen 6 complex, locus G6F HCG43720, isoform CRA_a; Lymphocyte antigen 6 complex locus protein G6f; Megakaryocyte-enhanced gene transcript 1 protein; Uncharacterized protein |
| chr20_-_36793663 | 0.11 |
ENST00000536701.1
ENST00000536724.1 |
TGM2
|
transglutaminase 2 |
| chr2_+_101591314 | 0.09 |
ENST00000450763.1
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr19_+_751122 | 0.08 |
ENST00000215582.6
|
MISP
|
mitotic spindle positioning |
| chr5_-_58571935 | 0.07 |
ENST00000503258.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr5_-_137548997 | 0.05 |
ENST00000505120.1
ENST00000394886.2 ENST00000394884.3 |
CDC23
|
cell division cycle 23 |
| chr9_+_33290491 | 0.01 |
ENST00000379540.3
ENST00000379521.4 ENST00000318524.6 |
NFX1
|
nuclear transcription factor, X-box binding 1 |
| chr15_+_69452959 | 0.00 |
ENST00000261858.2
|
GLCE
|
glucuronic acid epimerase |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.6 | 19.9 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 1.7 | 10.3 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.9 | 3.5 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.9 | 4.3 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.7 | 2.7 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.7 | 4.6 | GO:0032596 | protein transport within lipid bilayer(GO:0032594) protein transport into membrane raft(GO:0032596) |
| 0.6 | 1.8 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.5 | 1.5 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.5 | 2.0 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.5 | 7.3 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.4 | 2.6 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.4 | 3.5 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.4 | 1.3 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.3 | 3.8 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.3 | 2.3 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.3 | 2.2 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.3 | 2.9 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.2 | 2.0 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.2 | 24.4 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.2 | 1.0 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.2 | 1.7 | GO:0009838 | abscission(GO:0009838) |
| 0.2 | 1.4 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.2 | 1.0 | GO:2000323 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.2 | 22.6 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.2 | 1.4 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.2 | 0.5 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.2 | 2.2 | GO:0010666 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.2 | 1.5 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.1 | 0.3 | GO:0070662 | mast cell proliferation(GO:0070662) |
| 0.1 | 2.2 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.1 | 1.4 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 0.4 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.1 | 6.2 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.1 | 0.5 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 | 3.4 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.4 | GO:0035623 | regulation of pronephros size(GO:0035565) renal glucose absorption(GO:0035623) |
| 0.1 | 2.7 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 1.9 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 2.4 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 0.7 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 8.8 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 2.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.7 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 13.0 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.1 | 0.6 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.5 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.1 | 0.4 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 1.1 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 0.4 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.1 | 0.6 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.1 | 3.4 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.1 | 1.5 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 2.3 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 3.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 1.3 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 0.5 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.2 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.0 | 0.7 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.1 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.0 | 1.4 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 3.6 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 1.2 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 0.9 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.7 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.9 | GO:0050672 | negative regulation of mononuclear cell proliferation(GO:0032945) negative regulation of lymphocyte proliferation(GO:0050672) |
| 0.0 | 0.8 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.0 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.8 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.5 | 2.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.5 | 2.0 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.3 | 22.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.3 | 2.2 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.3 | 21.7 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.3 | 1.5 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.2 | 2.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 1.9 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 1.0 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.2 | 2.0 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 1.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 2.9 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.1 | 1.7 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 2.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.7 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 1.7 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.9 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 24.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 2.2 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 6.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.5 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 1.1 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 4.4 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 4.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.4 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.0 | 12.9 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.4 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 3.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 2.2 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 4.3 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 1.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.6 | 19.9 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 1.4 | 4.3 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 1.0 | 7.3 | GO:0004459 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.8 | 24.8 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.6 | 2.2 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.5 | 8.8 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.4 | 8.8 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.4 | 1.1 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.3 | 3.5 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.3 | 1.2 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.3 | 3.5 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.3 | 8.2 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.3 | 3.8 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.3 | 1.0 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.3 | 1.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 1.4 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.2 | 2.7 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.2 | 0.7 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.2 | 2.9 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 1.8 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 2.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.7 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.4 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 1.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 2.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 3.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 2.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 1.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.4 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.1 | 2.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 4.6 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 1.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 1.3 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 2.3 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.9 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 2.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.7 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.5 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 2.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 4.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.3 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.5 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 2.0 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 7.7 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.5 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.2 | 12.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 10.8 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 4.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 2.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 2.9 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 2.2 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 2.3 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 3.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 2.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.2 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.4 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 1.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 8.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.6 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 2.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 9.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.3 | 7.3 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.2 | 8.8 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.2 | 19.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.2 | 2.6 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 3.4 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.1 | 2.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 2.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 3.7 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 1.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 2.9 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.1 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 8.0 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 1.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 3.8 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 1.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 0.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 6.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 3.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.8 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 6.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 2.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 2.2 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.9 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.4 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |