Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for GLIS3
Z-value: 0.73
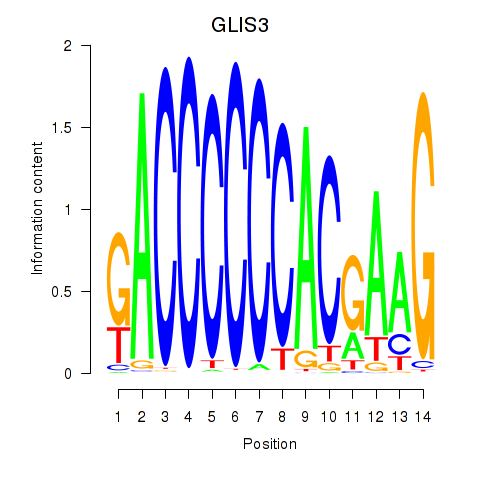
Transcription factors associated with GLIS3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GLIS3
|
ENSG00000107249.17 | GLIS family zinc finger 3 |
Activity profile of GLIS3 motif
Sorted Z-values of GLIS3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_48828582 | 17.95 |
ENST00000270221.6
ENST00000596315.1 |
EMP3
|
epithelial membrane protein 3 |
| chr18_+_3451584 | 9.60 |
ENST00000551541.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr1_-_45987526 | 9.01 |
ENST00000372079.1
ENST00000262746.1 ENST00000447184.1 ENST00000319248.8 |
PRDX1
|
peroxiredoxin 1 |
| chr1_-_45988542 | 8.87 |
ENST00000424390.1
|
PRDX1
|
peroxiredoxin 1 |
| chrX_+_64887512 | 8.76 |
ENST00000360270.5
|
MSN
|
moesin |
| chr19_-_41859814 | 7.89 |
ENST00000221930.5
|
TGFB1
|
transforming growth factor, beta 1 |
| chr21_-_46330545 | 7.63 |
ENST00000320216.6
ENST00000397852.1 |
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr5_+_172386419 | 7.11 |
ENST00000265100.2
ENST00000519239.1 |
RPL26L1
|
ribosomal protein L26-like 1 |
| chr7_+_150758304 | 6.87 |
ENST00000482950.1
ENST00000463414.1 ENST00000310317.5 |
SLC4A2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr5_-_180671172 | 6.75 |
ENST00000512805.1
|
GNB2L1
|
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 |
| chr18_+_3451646 | 6.62 |
ENST00000345133.5
ENST00000330513.5 ENST00000549546.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr3_-_25824925 | 5.52 |
ENST00000396649.3
ENST00000428257.1 ENST00000280700.5 |
NGLY1
|
N-glycanase 1 |
| chr11_+_68451943 | 5.51 |
ENST00000265643.3
|
GAL
|
galanin/GMAP prepropeptide |
| chr17_-_73178599 | 5.06 |
ENST00000578238.1
|
SUMO2
|
small ubiquitin-like modifier 2 |
| chr13_+_25670268 | 4.85 |
ENST00000281589.3
|
PABPC3
|
poly(A) binding protein, cytoplasmic 3 |
| chr3_-_25824872 | 4.41 |
ENST00000308710.5
|
NGLY1
|
N-glycanase 1 |
| chr6_+_44191507 | 4.11 |
ENST00000371724.1
ENST00000371713.1 |
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr3_+_38029462 | 3.78 |
ENST00000283713.6
|
VILL
|
villin-like |
| chr17_-_73179046 | 3.73 |
ENST00000314523.7
ENST00000420826.2 |
SUMO2
|
small ubiquitin-like modifier 2 |
| chr6_+_44191290 | 3.71 |
ENST00000371755.3
ENST00000371740.5 ENST00000371731.1 ENST00000393841.1 |
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr19_+_46850251 | 3.45 |
ENST00000012443.4
|
PPP5C
|
protein phosphatase 5, catalytic subunit |
| chr15_+_91478493 | 3.27 |
ENST00000418476.2
|
UNC45A
|
unc-45 homolog A (C. elegans) |
| chr11_-_8680383 | 3.24 |
ENST00000299550.6
|
TRIM66
|
tripartite motif containing 66 |
| chr19_+_46850320 | 3.20 |
ENST00000391919.1
|
PPP5C
|
protein phosphatase 5, catalytic subunit |
| chr4_-_84255935 | 3.10 |
ENST00000513463.1
|
HPSE
|
heparanase |
| chr1_-_42801540 | 3.06 |
ENST00000372573.1
|
FOXJ3
|
forkhead box J3 |
| chr6_+_33172407 | 3.03 |
ENST00000374662.3
|
HSD17B8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr22_-_50964849 | 2.99 |
ENST00000543927.1
ENST00000423348.1 |
SCO2
|
SCO2 cytochrome c oxidase assembly protein |
| chr6_+_89791507 | 2.76 |
ENST00000354922.3
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr14_+_22931924 | 2.67 |
ENST00000390477.2
|
TRDC
|
T cell receptor delta constant |
| chr22_+_50628999 | 2.56 |
ENST00000395827.1
|
TRABD
|
TraB domain containing |
| chr1_-_53018654 | 2.48 |
ENST00000257177.4
ENST00000355809.4 ENST00000528642.1 ENST00000470626.1 ENST00000371544.3 |
ZCCHC11
|
zinc finger, CCHC domain containing 11 |
| chr5_+_172386517 | 2.37 |
ENST00000519522.1
|
RPL26L1
|
ribosomal protein L26-like 1 |
| chr6_+_24495185 | 2.34 |
ENST00000348925.2
|
ALDH5A1
|
aldehyde dehydrogenase 5 family, member A1 |
| chr20_-_30310693 | 2.30 |
ENST00000307677.4
ENST00000420653.1 |
BCL2L1
|
BCL2-like 1 |
| chr22_+_31489344 | 2.25 |
ENST00000404574.1
|
SMTN
|
smoothelin |
| chr17_+_7792101 | 2.23 |
ENST00000358181.4
ENST00000330494.7 |
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chr20_-_30310656 | 2.03 |
ENST00000376055.4
|
BCL2L1
|
BCL2-like 1 |
| chr8_-_74791051 | 1.88 |
ENST00000453587.2
ENST00000602969.1 ENST00000602593.1 ENST00000419880.3 ENST00000517608.1 |
UBE2W
|
ubiquitin-conjugating enzyme E2W (putative) |
| chrX_-_132549506 | 1.83 |
ENST00000370828.3
|
GPC4
|
glypican 4 |
| chr10_+_105314881 | 1.53 |
ENST00000437579.1
|
NEURL
|
neuralized E3 ubiquitin protein ligase 1 |
| chr22_+_18834324 | 1.45 |
ENST00000342005.4
|
AC008132.13
|
Uncharacterized protein |
| chr2_-_208030647 | 1.40 |
ENST00000309446.6
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr1_-_42800860 | 1.21 |
ENST00000445886.1
ENST00000361346.1 ENST00000361776.1 |
FOXJ3
|
forkhead box J3 |
| chrX_+_118370288 | 1.16 |
ENST00000535419.1
|
PGRMC1
|
progesterone receptor membrane component 1 |
| chr19_+_41281060 | 1.11 |
ENST00000594436.1
ENST00000597784.1 |
MIA
|
melanoma inhibitory activity |
| chr17_-_3819751 | 0.99 |
ENST00000225538.3
|
P2RX1
|
purinergic receptor P2X, ligand-gated ion channel, 1 |
| chr18_+_11752040 | 0.92 |
ENST00000423027.3
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr17_-_5015129 | 0.89 |
ENST00000575898.1
ENST00000416429.2 |
ZNF232
|
zinc finger protein 232 |
| chr20_+_61867235 | 0.85 |
ENST00000342412.6
ENST00000217169.3 |
BIRC7
|
baculoviral IAP repeat containing 7 |
| chr18_+_11751493 | 0.85 |
ENST00000269162.5
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr6_+_24495067 | 0.83 |
ENST00000357578.3
ENST00000546278.1 ENST00000491546.1 |
ALDH5A1
|
aldehyde dehydrogenase 5 family, member A1 |
| chr19_-_10613421 | 0.82 |
ENST00000393623.2
|
KEAP1
|
kelch-like ECH-associated protein 1 |
| chrX_+_118370211 | 0.81 |
ENST00000217971.7
|
PGRMC1
|
progesterone receptor membrane component 1 |
| chr2_-_211168332 | 0.66 |
ENST00000341685.4
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast |
| chr3_+_173116225 | 0.62 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr11_+_66886717 | 0.50 |
ENST00000398645.2
|
KDM2A
|
lysine (K)-specific demethylase 2A |
| chr14_+_77228532 | 0.50 |
ENST00000167106.4
ENST00000554237.1 |
VASH1
|
vasohibin 1 |
| chr21_+_38071430 | 0.48 |
ENST00000290399.6
|
SIM2
|
single-minded family bHLH transcription factor 2 |
| chr12_+_132312931 | 0.45 |
ENST00000360564.1
ENST00000545671.1 ENST00000545790.1 |
MMP17
|
matrix metallopeptidase 17 (membrane-inserted) |
| chr20_+_18447771 | 0.38 |
ENST00000377603.4
|
POLR3F
|
polymerase (RNA) III (DNA directed) polypeptide F, 39 kDa |
| chr19_-_48673552 | 0.34 |
ENST00000536218.1
ENST00000596549.1 |
LIG1
|
ligase I, DNA, ATP-dependent |
| chr20_-_23969416 | 0.22 |
ENST00000335694.4
|
GGTLC1
|
gamma-glutamyltransferase light chain 1 |
| chr1_+_110527308 | 0.11 |
ENST00000369799.5
|
AHCYL1
|
adenosylhomocysteinase-like 1 |
| chr8_+_27183033 | 0.05 |
ENST00000420218.2
|
PTK2B
|
protein tyrosine kinase 2 beta |
Network of associatons between targets according to the STRING database.
First level regulatory network of GLIS3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.9 | GO:0052553 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 1.8 | 5.5 | GO:0051795 | cortisol secretion(GO:0043400) regulation of cortisol secretion(GO:0051462) positive regulation of catagen(GO:0051795) |
| 1.6 | 7.8 | GO:0015862 | uridine transport(GO:0015862) |
| 1.5 | 8.8 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 1.1 | 6.6 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.9 | 18.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.7 | 13.0 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.6 | 4.3 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.5 | 3.2 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.5 | 17.9 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.5 | 2.5 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.4 | 7.6 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.3 | 2.8 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.2 | 0.6 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.2 | 3.0 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.2 | 1.0 | GO:0002442 | serotonin production involved in inflammatory response(GO:0002351) serotonin secretion involved in inflammatory response(GO:0002442) serotonin secretion by platelet(GO:0002554) |
| 0.2 | 8.8 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.2 | 0.5 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 | 3.0 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) respiratory chain complex IV assembly(GO:0008535) |
| 0.1 | 6.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 9.5 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 0.8 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.3 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.1 | 3.8 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 1.9 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.1 | 0.5 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.9 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.5 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.0 | 2.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 3.0 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 2.2 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.1 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.0 | 12.9 | GO:0042493 | response to drug(GO:0042493) |
| 0.0 | 1.8 | GO:0001523 | retinoid metabolic process(GO:0001523) |
| 0.0 | 1.1 | GO:0008283 | cell proliferation(GO:0008283) |
| 0.0 | 2.3 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.0 | 0.2 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.4 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 7.6 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.9 | 6.6 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.4 | 8.8 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.3 | 4.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.5 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.1 | 17.9 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 9.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 2.2 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 9.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 3.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 8.8 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 2.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 1.0 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 14.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 3.1 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 2.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.7 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 9.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 2.0 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 5.5 | GO:0043025 | neuronal cell body(GO:0043025) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.9 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 2.2 | 17.9 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 1.4 | 5.5 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 1.1 | 7.6 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.8 | 3.1 | GO:0030305 | beta-glucuronidase activity(GO:0004566) heparanase activity(GO:0030305) |
| 0.7 | 8.8 | GO:0031386 | protein tag(GO:0031386) |
| 0.6 | 2.5 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.6 | 3.0 | GO:0070404 | NADH binding(GO:0070404) |
| 0.6 | 16.2 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.4 | 4.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.4 | 6.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.4 | 7.8 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.3 | 1.8 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.3 | 3.0 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.2 | 6.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 4.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 12.2 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 1.0 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 8.8 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 3.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 0.5 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 3.2 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 2.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 9.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.3 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 2.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 3.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.4 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.0 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.2 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 1.6 | GO:0005496 | steroid binding(GO:0005496) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 17.9 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.3 | 15.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 4.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 8.3 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.2 | 6.8 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 14.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 14.4 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 3.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 10.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.5 | PID NOTCH PATHWAY | Notch signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.3 | 16.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 8.8 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.2 | 4.9 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.2 | 7.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 4.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 3.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 1.0 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 7.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 9.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 6.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 1.5 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.3 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.0 | 3.5 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 2.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |