Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
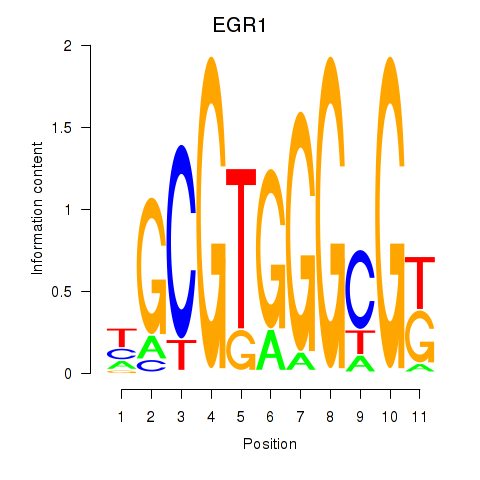
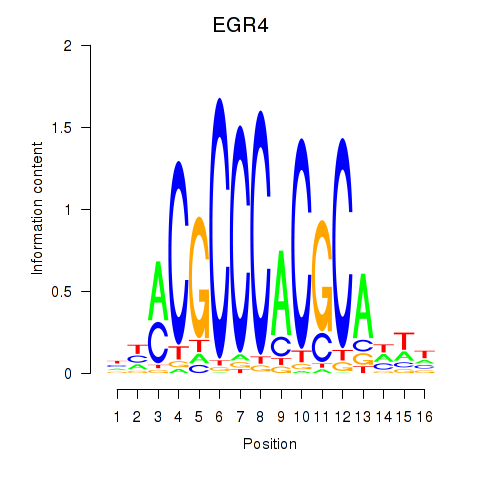
Results for EGR1_EGR4
Z-value: 0.54
Transcription factors associated with EGR1_EGR4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EGR1
|
ENSG00000120738.7 | early growth response 1 |
|
EGR4
|
ENSG00000135625.6 | early growth response 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EGR1 | hg19_v2_chr5_+_137801160_137801179 | 0.36 | 4.5e-08 | Click! |
| EGR4 | hg19_v2_chr2_-_73520667_73520833 | 0.18 | 8.2e-03 | Click! |
Activity profile of EGR1_EGR4 motif
Sorted Z-values of EGR1_EGR4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of EGR1_EGR4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.5 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 4.1 | 12.2 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 3.9 | 11.7 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 3.8 | 11.4 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 3.2 | 12.8 | GO:0001743 | optic placode formation(GO:0001743) |
| 2.5 | 7.5 | GO:1990922 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 2.4 | 7.2 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 2.1 | 8.3 | GO:0072237 | metanephric proximal tubule development(GO:0072237) |
| 1.9 | 5.7 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 1.6 | 4.9 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 1.6 | 6.5 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 1.6 | 25.7 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 1.6 | 4.8 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 1.5 | 3.0 | GO:0070434 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 1.4 | 2.9 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 1.4 | 4.2 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 1.4 | 8.2 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 1.4 | 5.4 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) regulation of polynucleotide adenylyltransferase activity(GO:1904245) |
| 1.3 | 3.8 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 1.3 | 6.3 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 1.2 | 3.7 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) negative regulation of osteoclast proliferation(GO:0090291) |
| 1.2 | 6.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 1.2 | 4.9 | GO:0003335 | corneocyte development(GO:0003335) |
| 1.2 | 3.6 | GO:0051595 | response to methylglyoxal(GO:0051595) |
| 1.2 | 9.5 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 1.2 | 4.7 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 1.1 | 5.7 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 1.1 | 3.4 | GO:0071529 | cementum mineralization(GO:0071529) |
| 1.1 | 5.5 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 1.1 | 6.5 | GO:1900239 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 1.1 | 6.4 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 1.0 | 4.2 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 1.0 | 2.0 | GO:0072014 | proximal tubule development(GO:0072014) |
| 1.0 | 3.0 | GO:0045994 | positive regulation of translational initiation by iron(GO:0045994) |
| 1.0 | 2.9 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 1.0 | 2.0 | GO:1905064 | negative regulation of vascular smooth muscle cell differentiation(GO:1905064) |
| 1.0 | 2.9 | GO:0051697 | protein delipidation(GO:0051697) |
| 1.0 | 9.6 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.9 | 2.8 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.9 | 7.5 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.9 | 2.8 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.9 | 1.8 | GO:0002537 | nitric oxide production involved in inflammatory response(GO:0002537) |
| 0.9 | 2.8 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.9 | 12.7 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.9 | 10.0 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.9 | 1.8 | GO:0051085 | 'de novo' posttranslational protein folding(GO:0051084) chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.9 | 4.5 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.9 | 3.5 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.9 | 5.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.9 | 5.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.9 | 2.6 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.9 | 1.7 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.8 | 9.3 | GO:0000050 | urea cycle(GO:0000050) |
| 0.8 | 2.5 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.8 | 2.5 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.8 | 5.0 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.8 | 0.8 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.8 | 2.4 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.8 | 10.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.8 | 2.3 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.8 | 2.3 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.8 | 3.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.8 | 6.1 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.8 | 1.5 | GO:1903521 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.8 | 1.5 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.7 | 10.2 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.7 | 20.1 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.7 | 2.1 | GO:0070105 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.7 | 9.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.7 | 2.8 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.7 | 2.1 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.7 | 1.4 | GO:0048382 | mesendoderm development(GO:0048382) |
| 0.7 | 2.0 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.7 | 6.0 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.7 | 2.6 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.7 | 0.7 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.6 | 5.2 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.6 | 2.6 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.6 | 1.9 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.6 | 1.3 | GO:2000705 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.6 | 3.2 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) response to odorant(GO:1990834) |
| 0.6 | 1.9 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.6 | 1.8 | GO:0010957 | negative regulation of vitamin D biosynthetic process(GO:0010957) |
| 0.6 | 3.7 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.6 | 1.7 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.6 | 3.4 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.6 | 1.7 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.6 | 1.7 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.6 | 3.9 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.5 | 3.3 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.5 | 1.6 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.5 | 4.3 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.5 | 1.6 | GO:1990737 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.5 | 3.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.5 | 2.1 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.5 | 2.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.5 | 2.0 | GO:0042418 | epinephrine biosynthetic process(GO:0042418) |
| 0.5 | 3.6 | GO:0050893 | sensory processing(GO:0050893) |
| 0.5 | 4.0 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.5 | 2.0 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.5 | 3.0 | GO:0034128 | negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) |
| 0.5 | 1.5 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.5 | 1.0 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.5 | 3.4 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.5 | 2.5 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.5 | 2.4 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 0.5 | 2.4 | GO:0021586 | pons maturation(GO:0021586) |
| 0.5 | 1.4 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.5 | 8.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.5 | 0.5 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.5 | 1.9 | GO:1903960 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) |
| 0.5 | 5.2 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.5 | 40.3 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.5 | 0.9 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.5 | 2.3 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.5 | 1.4 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.5 | 9.1 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.5 | 1.8 | GO:0072166 | posterior mesonephric tubule development(GO:0072166) negative regulation of metanephric glomerulus development(GO:0072299) regulation of metanephric glomerular mesangial cell proliferation(GO:0072301) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.5 | 0.5 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.5 | 5.0 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.4 | 1.8 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.4 | 2.2 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.4 | 1.3 | GO:0003342 | proepicardium development(GO:0003342) septum transversum development(GO:0003343) |
| 0.4 | 3.9 | GO:1902949 | positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.4 | 7.3 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.4 | 3.8 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.4 | 1.3 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.4 | 1.7 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.4 | 2.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.4 | 1.2 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.4 | 1.7 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.4 | 5.3 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.4 | 1.2 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.4 | 1.6 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.4 | 1.6 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.4 | 4.3 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.4 | 1.2 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.4 | 2.7 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.4 | 1.2 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.4 | 1.1 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.4 | 0.4 | GO:0085020 | protein K29-linked ubiquitination(GO:0035519) protein K27-linked ubiquitination(GO:0044314) protein K6-linked ubiquitination(GO:0085020) |
| 0.4 | 1.1 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.4 | 1.9 | GO:1901098 | regulation of autophagosome maturation(GO:1901096) positive regulation of autophagosome maturation(GO:1901098) |
| 0.4 | 2.6 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.4 | 1.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.4 | 1.5 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.4 | 4.4 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.4 | 6.1 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.4 | 4.3 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.3 | 3.5 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.3 | 0.7 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.3 | 3.4 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.3 | 1.4 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) ERK5 cascade(GO:0070375) |
| 0.3 | 1.0 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.3 | 2.3 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.3 | 2.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.3 | 10.4 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.3 | 12.0 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.3 | 3.6 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.3 | 4.3 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.3 | 1.3 | GO:1900107 | regulation of nodal signaling pathway(GO:1900107) |
| 0.3 | 2.0 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.3 | 1.0 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.3 | 1.3 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.3 | 1.0 | GO:0034184 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.3 | 5.1 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.3 | 1.6 | GO:0003431 | growth plate cartilage chondrocyte growth(GO:0003430) growth plate cartilage chondrocyte development(GO:0003431) Harderian gland development(GO:0070384) |
| 0.3 | 1.3 | GO:0046013 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.3 | 1.6 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.3 | 0.6 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.3 | 3.1 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.3 | 5.0 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.3 | 1.5 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.3 | 1.2 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.3 | 1.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.3 | 1.8 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.3 | 1.5 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.3 | 1.5 | GO:0072021 | ascending thin limb development(GO:0072021) metanephric ascending thin limb development(GO:0072218) |
| 0.3 | 2.1 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.3 | 3.6 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.3 | 1.8 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.3 | 0.9 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.3 | 9.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.3 | 9.0 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.3 | 2.3 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.3 | 1.4 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.3 | 2.9 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.3 | 5.2 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.3 | 4.0 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.3 | 2.6 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.3 | 1.1 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.3 | 7.6 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.3 | 1.1 | GO:1900738 | regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.3 | 2.5 | GO:1901998 | toxin transport(GO:1901998) |
| 0.3 | 1.7 | GO:0072103 | glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.3 | 2.2 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.3 | 1.1 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.3 | 2.2 | GO:0002326 | B cell lineage commitment(GO:0002326) immunoglobulin V(D)J recombination(GO:0033152) |
| 0.3 | 1.3 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.3 | 1.3 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.3 | 5.8 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.3 | 3.1 | GO:0007512 | adult heart development(GO:0007512) |
| 0.3 | 3.1 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.3 | 2.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.3 | 0.5 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.3 | 1.0 | GO:0009648 | photoperiodism(GO:0009648) entrainment of circadian clock(GO:0009649) entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.2 | 1.2 | GO:2000468 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) regulation of peroxidase activity(GO:2000468) positive regulation of peroxidase activity(GO:2000470) |
| 0.2 | 1.5 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.2 | 1.2 | GO:0070091 | glucagon secretion(GO:0070091) regulation of glucagon secretion(GO:0070092) |
| 0.2 | 0.5 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.2 | 1.4 | GO:0052551 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.2 | 2.1 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.2 | 3.7 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 1.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.2 | 5.0 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.2 | 1.8 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.2 | 13.1 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.2 | 2.4 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.2 | 3.5 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.2 | 0.2 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.2 | 2.5 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.2 | 0.6 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.2 | 1.9 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.2 | 0.8 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.2 | 2.9 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.2 | 0.4 | GO:0035900 | response to isolation stress(GO:0035900) |
| 0.2 | 6.4 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.2 | 3.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 2.2 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.2 | 3.0 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.2 | 1.0 | GO:0009137 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.2 | 2.8 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.2 | 1.6 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.2 | 2.6 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.2 | 3.4 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.2 | 0.6 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.2 | 4.9 | GO:0097503 | sialylation(GO:0097503) |
| 0.2 | 0.8 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.2 | 4.9 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.2 | 0.9 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.2 | 1.1 | GO:0043320 | natural killer cell degranulation(GO:0043320) |
| 0.2 | 1.1 | GO:0036289 | peptidyl-serine autophosphorylation(GO:0036289) |
| 0.2 | 7.5 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.2 | 1.7 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.2 | 0.6 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.2 | 0.5 | GO:0032954 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.2 | 6.4 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.2 | 2.2 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.2 | 2.7 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.2 | 2.5 | GO:0033127 | regulation of histone phosphorylation(GO:0033127) |
| 0.2 | 1.6 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.2 | 0.9 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.2 | 1.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.2 | 2.4 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.2 | 2.4 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.2 | 1.7 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.2 | 6.6 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.2 | 0.8 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.2 | 4.3 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.2 | 0.7 | GO:0060084 | positive regulation of transmission of nerve impulse(GO:0051971) synaptic transmission involved in micturition(GO:0060084) |
| 0.2 | 3.9 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.2 | 3.4 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.2 | 3.8 | GO:0097502 | mannosylation(GO:0097502) |
| 0.2 | 1.9 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.2 | 0.6 | GO:1901184 | regulation of ERBB signaling pathway(GO:1901184) |
| 0.2 | 0.8 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.2 | 1.1 | GO:0032097 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.1 | 6.1 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 1.9 | GO:0007034 | vacuolar transport(GO:0007034) |
| 0.1 | 2.3 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.1 | 0.4 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.1 | 1.0 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.7 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.1 | 1.4 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 1.4 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 1.5 | GO:0006024 | aminoglycan biosynthetic process(GO:0006023) glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.1 | 1.9 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.1 | 1.5 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.1 | 0.9 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.1 | 0.3 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 0.7 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.1 | 0.7 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 1.2 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.1 | 1.1 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.1 | 0.7 | GO:0046959 | habituation(GO:0046959) |
| 0.1 | 0.8 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 2.1 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 2.6 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.1 | 1.1 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 2.0 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 1.0 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.2 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.1 | 0.5 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 1.4 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.1 | 0.7 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 2.5 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.1 | 1.4 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.1 | 1.0 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 1.4 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 2.5 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.1 | 0.7 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 3.6 | GO:0001510 | RNA methylation(GO:0001510) |
| 0.1 | 1.4 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.1 | 0.8 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.2 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.1 | 0.4 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 0.8 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.1 | 0.7 | GO:0042711 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.1 | 2.5 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 1.6 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 2.3 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.1 | 0.2 | GO:0044065 | regulation of respiratory gaseous exchange(GO:0043576) regulation of respiratory system process(GO:0044065) |
| 0.1 | 1.1 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.1 | 0.8 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 5.0 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.1 | 0.6 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 1.4 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.1 | 1.3 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.1 | 3.9 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 0.3 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.1 | 0.5 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.1 | 0.5 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 1.9 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 1.6 | GO:0030324 | lung development(GO:0030324) |
| 0.1 | 1.6 | GO:0043243 | positive regulation of protein complex disassembly(GO:0043243) |
| 0.1 | 0.9 | GO:0031124 | mRNA 3'-end processing(GO:0031124) RNA polyadenylation(GO:0043631) |
| 0.1 | 0.8 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.1 | 1.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.3 | GO:2001140 | regulation of phospholipid transport(GO:2001138) positive regulation of phospholipid transport(GO:2001140) |
| 0.1 | 0.5 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.1 | 0.5 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.1 | 2.7 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 | 4.3 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.1 | 0.6 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.3 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 1.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.8 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.1 | 0.8 | GO:0023019 | signal transduction involved in regulation of gene expression(GO:0023019) |
| 0.1 | 0.4 | GO:1901911 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 1.4 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.1 | 1.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.7 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.1 | 1.6 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.1 | 0.6 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.1 | 0.6 | GO:0097107 | postsynaptic density organization(GO:0097106) postsynaptic density assembly(GO:0097107) gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.1 | 0.6 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.1 | 2.1 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.1 | 0.2 | GO:1901166 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.1 | 0.8 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 3.3 | GO:0010830 | regulation of myotube differentiation(GO:0010830) |
| 0.1 | 0.3 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.1 | 0.7 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.1 | 1.0 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 1.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.3 | GO:0072141 | renal interstitial fibroblast development(GO:0072141) mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) |
| 0.1 | 0.4 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 | 1.1 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 0.4 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 2.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 3.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 0.7 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.1 | 1.0 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.1 | 12.5 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 2.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.5 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.1 | 0.5 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.1 | 2.1 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.1 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.5 | GO:0006349 | regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.1 | 0.3 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 0.3 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.4 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.1 | 0.2 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.1 | 0.4 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.1 | 0.5 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.1 | 0.4 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.1 | 1.4 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 0.7 | GO:2000696 | regulation of epithelial cell differentiation involved in kidney development(GO:2000696) |
| 0.1 | 0.7 | GO:0043586 | tongue development(GO:0043586) |
| 0.1 | 0.1 | GO:0019860 | uracil metabolic process(GO:0019860) |
| 0.1 | 0.4 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.1 | 0.7 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.7 | GO:0003299 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.1 | 0.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 4.5 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.1 | 3.9 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 0.8 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.1 | 2.0 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 0.6 | GO:1903861 | regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 1.6 | GO:0048512 | circadian behavior(GO:0048512) |
| 0.1 | 0.9 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 1.3 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.5 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.2 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.0 | 1.7 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.0 | 0.7 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.9 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.7 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 1.5 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 6.3 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 1.3 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 1.0 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.5 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.8 | GO:0007140 | male meiosis(GO:0007140) |
| 0.0 | 0.4 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.9 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.9 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.7 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.0 | 0.3 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 0.1 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.0 | 1.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 1.2 | GO:0035303 | regulation of dephosphorylation(GO:0035303) |
| 0.0 | 0.3 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 1.0 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.3 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.0 | 0.3 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.7 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.4 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.0 | 0.2 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 1.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 1.1 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.4 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.2 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 0.3 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.5 | GO:0048485 | sympathetic nervous system development(GO:0048485) |
| 0.0 | 0.6 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 1.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.0 | 0.4 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.7 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0070458 | cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 | 1.3 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 1.0 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.0 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 1.4 | GO:0050709 | negative regulation of protein secretion(GO:0050709) |
| 0.0 | 0.1 | GO:1901142 | insulin processing(GO:0030070) insulin metabolic process(GO:1901142) |
| 0.0 | 0.6 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.7 | GO:0030218 | erythrocyte differentiation(GO:0030218) |
| 0.0 | 1.5 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.0 | 0.1 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.3 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.2 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.9 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.6 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 2.2 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.2 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 2.4 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.2 | GO:0000394 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) |
| 0.0 | 0.2 | GO:0019054 | modulation by virus of host process(GO:0019054) |
| 0.0 | 1.1 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.0 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 1.0 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.0 | 0.2 | GO:2000505 | regulation of energy homeostasis(GO:2000505) |
| 0.0 | 0.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.2 | GO:2000177 | regulation of neural precursor cell proliferation(GO:2000177) |
| 0.0 | 0.1 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.5 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.2 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 1.0 | GO:0098869 | cellular oxidant detoxification(GO:0098869) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0008356 | asymmetric cell division(GO:0008356) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 7.6 | GO:0045298 | tubulin complex(GO:0045298) |
| 1.3 | 5.2 | GO:0043293 | apoptosome(GO:0043293) |
| 1.1 | 5.6 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 1.0 | 6.1 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.8 | 3.1 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.8 | 3.0 | GO:0031417 | NatC complex(GO:0031417) |
| 0.7 | 10.9 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.7 | 2.2 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.7 | 2.2 | GO:0000806 | Y chromosome(GO:0000806) |
| 0.7 | 2.1 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.6 | 2.6 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.6 | 3.8 | GO:0000805 | X chromosome(GO:0000805) Barr body(GO:0001740) |
| 0.6 | 3.7 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.6 | 4.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.6 | 3.4 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.6 | 8.4 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.5 | 6.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.5 | 11.8 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.5 | 3.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.5 | 2.0 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.5 | 4.7 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.5 | 4.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.4 | 1.3 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.4 | 1.3 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.4 | 7.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.4 | 1.7 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.4 | 1.3 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.4 | 2.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.4 | 5.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.4 | 5.4 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.4 | 3.8 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.4 | 7.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.4 | 6.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.4 | 2.5 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.4 | 2.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.3 | 1.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.3 | 4.0 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.3 | 5.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.3 | 2.8 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.3 | 1.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.3 | 0.9 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.3 | 2.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.3 | 1.5 | GO:0032021 | NELF complex(GO:0032021) |
| 0.3 | 7.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.3 | 39.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.3 | 1.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.3 | 2.5 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.3 | 12.8 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.3 | 7.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.3 | 1.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.3 | 14.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.3 | 3.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.3 | 0.8 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.3 | 1.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.2 | 0.7 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.2 | 1.0 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.2 | 2.6 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.2 | 25.9 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.2 | 0.7 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.2 | 1.4 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.2 | 0.7 | GO:0031213 | RSF complex(GO:0031213) |
| 0.2 | 1.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 1.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.2 | 1.1 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.2 | 1.7 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.2 | 0.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.2 | 3.9 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.2 | 1.3 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.2 | 1.1 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.2 | 1.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.2 | 1.6 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.2 | 0.7 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.2 | 2.1 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.2 | 2.6 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.2 | 2.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 4.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 4.0 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.2 | 2.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.2 | 2.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 1.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.2 | 1.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 5.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 2.0 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 2.9 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 1.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 7.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 1.1 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.1 | 3.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 2.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.9 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 4.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 4.2 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 1.6 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 1.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.1 | 1.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 6.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 1.2 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 3.0 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.1 | 1.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 2.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 10.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 1.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 5.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 1.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 0.5 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 1.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 5.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.9 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.5 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 7.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 0.7 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 9.8 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 1.8 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 0.4 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 1.0 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.5 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 6.2 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 0.5 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.1 | 2.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 25.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 1.6 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 1.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 2.6 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 0.6 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.9 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 4.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 0.6 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 22.4 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 3.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 14.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 2.6 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 2.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.7 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 1.0 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 1.5 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 2.1 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 3.4 | GO:0042175 | nuclear outer membrane-endoplasmic reticulum membrane network(GO:0042175) |
| 0.0 | 1.2 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 3.0 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 2.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 2.2 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.3 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 1.5 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 3.1 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 1.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.2 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 1.7 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.1 | GO:0044292 | dendrite terminus(GO:0044292) dendritic growth cone(GO:0044294) |
| 0.0 | 1.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.2 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.0 | 1.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 3.3 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 0.5 | GO:0044297 | cell body(GO:0044297) |
| 0.0 | 0.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 2.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 16.1 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 4.2 | 12.5 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 3.8 | 11.4 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 3.7 | 11.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 3.1 | 9.2 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 2.5 | 7.6 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 2.1 | 37.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 2.1 | 8.2 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 2.0 | 6.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 1.5 | 15.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 1.5 | 7.5 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 1.4 | 7.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 1.4 | 7.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 1.3 | 4.0 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 1.2 | 6.1 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 1.1 | 3.2 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) |
| 1.0 | 10.2 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 1.0 | 13.9 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 1.0 | 4.0 | GO:0035939 | microsatellite binding(GO:0035939) |
| 1.0 | 1.9 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 1.0 | 3.8 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.9 | 12.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.9 | 12.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.9 | 3.5 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.9 | 2.6 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.9 | 4.3 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.8 | 1.6 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.8 | 10.2 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.8 | 3.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.8 | 2.3 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.7 | 4.3 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.7 | 2.1 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.7 | 9.6 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.7 | 7.5 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.7 | 1.4 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.7 | 2.0 | GO:0004603 | phenylethanolamine N-methyltransferase activity(GO:0004603) |
| 0.7 | 2.0 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.6 | 3.8 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.6 | 15.2 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.6 | 1.8 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.6 | 2.4 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.6 | 3.7 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.6 | 1.8 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.6 | 3.4 | GO:0004673 | phosphorelay sensor kinase activity(GO:0000155) protein histidine kinase activity(GO:0004673) |
| 0.6 | 2.3 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.6 | 1.7 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.6 | 1.7 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.6 | 1.7 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.5 | 3.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.5 | 4.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.5 | 1.6 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.5 | 1.6 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.5 | 1.6 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.5 | 1.6 | GO:0035473 | lipase binding(GO:0035473) |
| 0.5 | 7.8 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.5 | 1.6 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.5 | 14.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.5 | 2.0 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.5 | 3.4 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.5 | 7.7 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.5 | 6.7 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.5 | 4.7 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.5 | 2.8 | GO:0008430 | selenium binding(GO:0008430) |
| 0.5 | 4.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.5 | 2.7 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.4 | 1.7 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.4 | 19.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.4 | 1.3 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.4 | 3.0 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.4 | 1.3 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.4 | 2.5 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.4 | 5.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.4 | 8.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.4 | 6.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.4 | 1.6 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.4 | 1.2 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.4 | 2.3 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.4 | 1.5 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.4 | 14.0 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.4 | 4.9 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.4 | 1.8 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.4 | 2.8 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.3 | 5.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.3 | 1.7 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.3 | 2.7 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.3 | 3.8 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.3 | 1.6 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.3 | 2.5 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.3 | 4.8 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.3 | 10.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.3 | 1.4 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.3 | 3.6 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.3 | 2.8 | GO:0004075 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.3 | 0.8 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.3 | 2.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.3 | 1.9 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.3 | 1.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.3 | 1.3 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.3 | 3.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.3 | 2.1 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.3 | 5.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.3 | 1.5 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.3 | 0.8 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.3 | 1.3 | GO:0070404 | NADH binding(GO:0070404) |
| 0.3 | 2.5 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 1.2 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.2 | 20.1 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.2 | 1.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 0.7 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.2 | 2.8 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.2 | 0.7 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.2 | 1.1 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.2 | 3.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 1.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.2 | 0.6 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.2 | 1.3 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.2 | 4.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.2 | 0.6 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.2 | 7.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.2 | 2.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.2 | 1.5 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.2 | 7.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.2 | 4.2 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.2 | 4.1 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.2 | 7.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.2 | 12.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.2 | 1.7 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.2 | 1.7 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.2 | 1.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 0.9 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 2.0 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.2 | 4.6 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.2 | 2.0 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.2 | 0.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.2 | 2.9 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 1.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.2 | 1.4 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.2 | 3.9 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.2 | 1.6 | GO:0004691 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 1.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 2.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 0.3 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.2 | 5.9 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.2 | 11.4 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.2 | 1.0 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.2 | 3.7 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.2 | 6.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.2 | 1.2 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.2 | 8.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 1.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 5.7 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 0.4 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.1 | 17.3 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 3.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 1.1 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.1 | 0.4 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 1.9 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 2.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 1.8 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.8 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.5 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 2.8 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.5 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 1.0 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 6.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 1.0 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 1.5 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 1.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.5 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 0.5 | GO:0004566 | beta-glucuronidase activity(GO:0004566) beta-glucosidase activity(GO:0008422) |
| 0.1 | 2.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 3.4 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 2.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.6 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 1.6 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 1.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 10.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 0.9 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.1 | 1.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.7 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 1.2 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.1 | 1.8 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.3 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.1 | 0.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 1.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 0.5 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 0.3 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 1.6 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 3.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.6 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.1 | 3.6 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.1 | 1.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.9 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 2.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 0.4 | GO:0052843 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.1 | 0.3 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 3.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 1.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 2.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.7 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.1 | 4.3 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 0.4 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 1.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.5 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.7 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 6.4 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 8.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 3.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 25.6 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.1 | 1.0 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 0.5 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 3.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 5.3 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 1.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.6 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.1 | 0.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 1.1 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.9 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.2 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 1.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 28.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 1.0 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 1.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.6 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 0.2 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.1 | 1.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.3 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.1 | 1.7 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 1.9 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 1.1 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 4.4 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.1 | 1.4 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.1 | 1.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.5 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 2.0 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 0.3 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 2.6 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 0.3 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 0.3 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.1 | 1.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 3.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.7 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 1.0 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 10.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 1.2 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 2.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 2.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.5 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.3 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 2.3 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.8 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 1.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 1.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.7 | GO:0042165 | acetylcholine-gated cation channel activity(GO:0022848) neurotransmitter binding(GO:0042165) acetylcholine binding(GO:0042166) |
| 0.0 | 0.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 2.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 1.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 19.7 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 1.9 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
| 0.0 | 0.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 1.2 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.0 | GO:0051765 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol tetrakisphosphate kinase activity(GO:0051765) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.5 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 1.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 52.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.7 | 20.5 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.7 | 8.5 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.6 | 19.9 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.6 | 11.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.6 | 29.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.6 | 6.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.5 | 19.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.4 | 3.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.4 | 6.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.3 | 5.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.3 | 3.6 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.3 | 3.9 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.3 | 13.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.3 | 21.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.3 | 3.8 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.3 | 1.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.3 | 1.6 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.3 | 7.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 5.2 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.2 | 6.9 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.2 | 4.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.2 | 8.5 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.2 | 13.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 11.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.2 | 8.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 1.2 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.2 | 5.7 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.2 | 7.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.2 | 6.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.2 | 3.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 1.7 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.2 | 7.1 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.2 | 5.9 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 1.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 8.4 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 6.8 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 8.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 1.5 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.1 | 0.5 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 5.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 0.7 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 1.1 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 2.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 2.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 0.9 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 0.7 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 1.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 5.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 3.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 1.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 1.8 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 2.1 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 2.3 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 2.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 0.5 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 2.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 2.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.6 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 1.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 2.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.5 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.9 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.4 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.1 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 1.7 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 1.3 | 3.9 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.9 | 55.8 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.9 | 15.5 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.9 | 4.3 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.6 | 0.6 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.6 | 6.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.6 | 13.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.5 | 10.8 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.5 | 9.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.5 | 5.7 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.5 | 18.9 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.4 | 7.8 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.4 | 1.3 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.4 | 9.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.4 | 1.4 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.3 | 5.5 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.3 | 8.8 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.3 | 1.2 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.3 | 2.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.3 | 5.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.3 | 3.4 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.3 | 4.8 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.2 | 8.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.2 | 5.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 4.2 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.2 | 4.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.2 | 10.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 3.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 18.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.2 | 4.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 5.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.2 | 3.5 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.2 | 6.3 | REACTOME NEGATIVE REGULATION OF FGFR SIGNALING | Genes involved in Negative regulation of FGFR signaling |
| 0.2 | 6.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 5.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.2 | 1.7 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.2 | 2.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.2 | 7.5 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.2 | 4.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 4.3 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.2 | 3.0 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 1.7 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.2 | 5.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 3.9 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 3.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 0.4 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 2.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 4.3 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 2.7 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 1.2 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.1 | 8.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 1.9 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 15.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 1.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 1.4 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 2.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 4.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 0.8 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 0.9 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 1.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 2.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 2.3 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 3.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.9 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 1.4 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 2.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 0.6 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 1.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 2.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 2.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 4.2 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 6.3 | REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | Genes involved in RNA Polymerase II Pre-transcription Events |
| 0.1 | 1.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.1 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.1 | 2.9 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 1.8 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 1.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 3.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 3.1 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 1.2 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 1.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 0.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.0 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.1 | 1.0 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 8.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 3.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.8 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 2.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.8 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.7 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 1.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.5 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.8 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 2.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |