Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for BCL6B
Z-value: 0.09
Transcription factors associated with BCL6B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BCL6B
|
ENSG00000161940.6 | BCL6B transcription repressor |
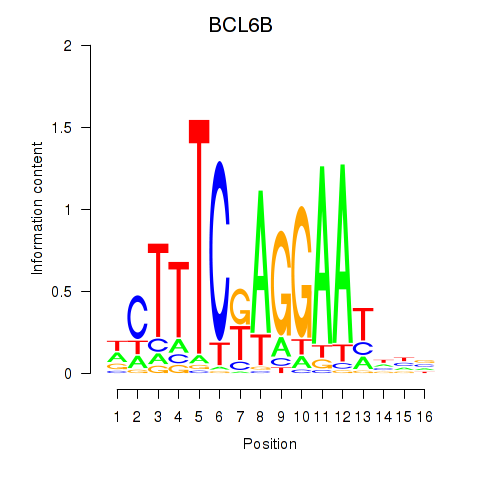
Activity profile of BCL6B motif
Sorted Z-values of BCL6B motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_104480019 | 6.61 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr12_-_10007448 | 5.69 |
ENST00000538152.1
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr12_+_104324112 | 4.44 |
ENST00000299767.5
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr5_-_172198190 | 4.01 |
ENST00000239223.3
|
DUSP1
|
dual specificity phosphatase 1 |
| chr16_-_75498308 | 3.60 |
ENST00000569540.1
|
TMEM170A
|
transmembrane protein 170A |
| chr16_+_58283814 | 3.20 |
ENST00000443128.2
ENST00000219299.4 |
CCDC113
|
coiled-coil domain containing 113 |
| chr1_-_108735440 | 2.82 |
ENST00000370041.4
|
SLC25A24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr3_+_23958470 | 2.54 |
ENST00000434031.2
ENST00000413699.1 ENST00000456530.2 |
RPL15
|
ribosomal protein L15 |
| chr6_+_42584847 | 2.53 |
ENST00000372883.3
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr5_+_54398463 | 2.46 |
ENST00000274306.6
|
GZMA
|
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3) |
| chr11_+_58912240 | 2.38 |
ENST00000527629.1
ENST00000361723.3 ENST00000531408.1 |
FAM111A
|
family with sequence similarity 111, member A |
| chr3_-_123512688 | 2.34 |
ENST00000475616.1
|
MYLK
|
myosin light chain kinase |
| chr1_+_158815588 | 2.30 |
ENST00000438394.1
|
MNDA
|
myeloid cell nuclear differentiation antigen |
| chr10_+_11207438 | 2.24 |
ENST00000609692.1
ENST00000354897.3 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr15_+_45003675 | 2.23 |
ENST00000558401.1
ENST00000559916.1 ENST00000544417.1 |
B2M
|
beta-2-microglobulin |
| chr13_-_33112956 | 2.18 |
ENST00000505213.1
|
N4BP2L2
|
NEDD4 binding protein 2-like 2 |
| chr13_-_33112899 | 2.05 |
ENST00000267068.3
ENST00000357505.6 ENST00000399396.3 |
N4BP2L2
|
NEDD4 binding protein 2-like 2 |
| chr1_-_212004090 | 2.03 |
ENST00000366997.4
|
LPGAT1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr2_+_143635067 | 1.95 |
ENST00000264170.4
|
KYNU
|
kynureninase |
| chr12_-_91574142 | 1.95 |
ENST00000547937.1
|
DCN
|
decorin |
| chr12_+_93964746 | 1.94 |
ENST00000536696.2
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr12_-_123201337 | 1.93 |
ENST00000528880.2
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr18_-_52989217 | 1.87 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr7_+_134464414 | 1.86 |
ENST00000361901.2
|
CALD1
|
caldesmon 1 |
| chr1_+_84609944 | 1.84 |
ENST00000370685.3
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr14_-_25103388 | 1.78 |
ENST00000526004.1
ENST00000415355.3 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr6_+_35995531 | 1.78 |
ENST00000229794.4
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr12_-_123187890 | 1.76 |
ENST00000328880.5
|
HCAR2
|
hydroxycarboxylic acid receptor 2 |
| chr18_-_52989525 | 1.70 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr12_-_120687948 | 1.68 |
ENST00000458477.2
|
PXN
|
paxillin |
| chr17_-_48474828 | 1.59 |
ENST00000576448.1
ENST00000225972.7 |
LRRC59
|
leucine rich repeat containing 59 |
| chr13_+_53191605 | 1.57 |
ENST00000342657.3
ENST00000398039.1 |
HNRNPA1L2
|
heterogeneous nuclear ribonucleoprotein A1-like 2 |
| chr19_+_55128576 | 1.55 |
ENST00000396331.1
|
LILRB1
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 1 |
| chr9_-_117880477 | 1.55 |
ENST00000534839.1
ENST00000340094.3 ENST00000535648.1 ENST00000346706.3 ENST00000345230.3 ENST00000350763.4 |
TNC
|
tenascin C |
| chr3_-_123603137 | 1.51 |
ENST00000360304.3
ENST00000359169.1 ENST00000346322.5 ENST00000360772.3 |
MYLK
|
myosin light chain kinase |
| chr14_-_25103472 | 1.43 |
ENST00000216341.4
ENST00000382542.1 ENST00000382540.1 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr7_-_29186008 | 1.41 |
ENST00000396276.3
ENST00000265394.5 |
CPVL
|
carboxypeptidase, vitellogenic-like |
| chr21_+_17102311 | 1.39 |
ENST00000285679.6
ENST00000351097.5 ENST00000285681.2 ENST00000400183.2 |
USP25
|
ubiquitin specific peptidase 25 |
| chr6_+_35995552 | 1.37 |
ENST00000468133.1
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr13_+_20207782 | 1.36 |
ENST00000414242.2
ENST00000361479.5 |
MPHOSPH8
|
M-phase phosphoprotein 8 |
| chr6_+_35995488 | 1.36 |
ENST00000229795.3
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr19_+_10197463 | 1.35 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr19_-_13227463 | 1.34 |
ENST00000437766.1
ENST00000221504.8 |
TRMT1
|
tRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr9_-_113018835 | 1.34 |
ENST00000374517.5
|
TXN
|
thioredoxin |
| chr2_-_216257849 | 1.28 |
ENST00000456923.1
|
FN1
|
fibronectin 1 |
| chr2_-_188419200 | 1.25 |
ENST00000233156.3
ENST00000426055.1 ENST00000453013.1 ENST00000417013.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr2_-_55237484 | 1.22 |
ENST00000394609.2
|
RTN4
|
reticulon 4 |
| chr1_-_150669604 | 1.21 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr19_-_13227534 | 1.21 |
ENST00000588229.1
ENST00000357720.4 |
TRMT1
|
tRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr14_+_75988851 | 1.19 |
ENST00000555504.1
|
BATF
|
basic leucine zipper transcription factor, ATF-like |
| chr2_+_70121075 | 1.15 |
ENST00000409116.1
|
SNRNP27
|
small nuclear ribonucleoprotein 27kDa (U4/U6.U5) |
| chr6_+_44194762 | 1.12 |
ENST00000371708.1
|
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr7_+_134464376 | 1.10 |
ENST00000454108.1
ENST00000361675.2 |
CALD1
|
caldesmon 1 |
| chr12_-_122985067 | 1.03 |
ENST00000540586.1
ENST00000543897.1 |
ZCCHC8
|
zinc finger, CCHC domain containing 8 |
| chr11_+_18230685 | 1.02 |
ENST00000340135.3
ENST00000534640.1 |
RP11-113D6.10
|
Putative mitochondrial carrier protein LOC494141 |
| chr2_+_143635222 | 1.01 |
ENST00000375773.2
ENST00000409512.1 ENST00000410015.2 |
KYNU
|
kynureninase |
| chr3_+_23959185 | 0.99 |
ENST00000354811.5
|
RPL15
|
ribosomal protein L15 |
| chr1_+_221051699 | 0.93 |
ENST00000366903.6
|
HLX
|
H2.0-like homeobox |
| chrX_+_153775821 | 0.88 |
ENST00000263518.6
ENST00000470142.1 ENST00000393549.2 ENST00000455588.2 ENST00000369602.3 |
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr12_-_122985494 | 0.88 |
ENST00000336229.4
|
ZCCHC8
|
zinc finger, CCHC domain containing 8 |
| chr13_+_73356197 | 0.87 |
ENST00000326291.6
|
PIBF1
|
progesterone immunomodulatory binding factor 1 |
| chr18_+_72167096 | 0.85 |
ENST00000324301.8
|
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr11_+_18230727 | 0.85 |
ENST00000527059.1
|
RP11-113D6.10
|
Putative mitochondrial carrier protein LOC494141 |
| chr3_+_50649302 | 0.79 |
ENST00000446044.1
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr3_+_23958632 | 0.78 |
ENST00000412097.1
ENST00000415719.1 ENST00000435882.1 |
RPL15
|
ribosomal protein L15 |
| chr6_+_150690133 | 0.78 |
ENST00000392255.3
ENST00000500320.3 |
IYD
|
iodotyrosine deiodinase |
| chr1_-_193029192 | 0.76 |
ENST00000417752.1
ENST00000367452.4 |
UCHL5
|
ubiquitin carboxyl-terminal hydrolase L5 |
| chr1_-_150669500 | 0.74 |
ENST00000271732.3
|
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr12_+_16064106 | 0.73 |
ENST00000428559.2
|
DERA
|
deoxyribose-phosphate aldolase (putative) |
| chr13_-_33112823 | 0.72 |
ENST00000504114.1
|
N4BP2L2
|
NEDD4 binding protein 2-like 2 |
| chr19_-_40023450 | 0.71 |
ENST00000326282.4
|
EID2B
|
EP300 interacting inhibitor of differentiation 2B |
| chr17_-_42907564 | 0.70 |
ENST00000592524.1
|
GJC1
|
gap junction protein, gamma 1, 45kDa |
| chr5_-_140700322 | 0.67 |
ENST00000313368.5
|
TAF7
|
TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 55kDa |
| chr12_+_19358228 | 0.65 |
ENST00000424268.1
ENST00000543806.1 |
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chrX_+_95939638 | 0.62 |
ENST00000373061.3
ENST00000373054.4 ENST00000355827.4 |
DIAPH2
|
diaphanous-related formin 2 |
| chrX_-_57147748 | 0.59 |
ENST00000374910.3
|
SPIN2B
|
spindlin family, member 2B |
| chr22_+_45725524 | 0.59 |
ENST00000405548.3
|
FAM118A
|
family with sequence similarity 118, member A |
| chr14_+_95078714 | 0.59 |
ENST00000393078.3
ENST00000393080.4 ENST00000467132.1 |
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr12_+_8975061 | 0.57 |
ENST00000299698.7
|
A2ML1
|
alpha-2-macroglobulin-like 1 |
| chr7_+_74072011 | 0.57 |
ENST00000324896.4
ENST00000353920.4 ENST00000346152.4 ENST00000416070.1 |
GTF2I
|
general transcription factor IIi |
| chr2_-_175499294 | 0.55 |
ENST00000392547.2
|
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr1_+_209878182 | 0.55 |
ENST00000367027.3
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr1_-_168698433 | 0.51 |
ENST00000367817.3
|
DPT
|
dermatopontin |
| chr11_+_123986069 | 0.50 |
ENST00000456829.2
ENST00000361352.5 ENST00000449321.1 ENST00000392748.1 ENST00000360334.4 ENST00000392744.4 |
VWA5A
|
von Willebrand factor A domain containing 5A |
| chr2_-_188419078 | 0.43 |
ENST00000437725.1
ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr11_+_113779704 | 0.42 |
ENST00000537778.1
|
HTR3B
|
5-hydroxytryptamine (serotonin) receptor 3B, ionotropic |
| chr10_-_70287172 | 0.40 |
ENST00000539557.1
|
SLC25A16
|
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
| chr19_+_45147313 | 0.38 |
ENST00000406449.4
|
PVR
|
poliovirus receptor |
| chr6_+_7541845 | 0.38 |
ENST00000418664.2
|
DSP
|
desmoplakin |
| chr3_+_158787041 | 0.37 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr17_+_38497640 | 0.35 |
ENST00000394086.3
|
RARA
|
retinoic acid receptor, alpha |
| chr3_-_69101461 | 0.34 |
ENST00000543976.1
|
TMF1
|
TATA element modulatory factor 1 |
| chr17_-_42908155 | 0.31 |
ENST00000426548.1
ENST00000590758.1 ENST00000591424.1 |
GJC1
|
gap junction protein, gamma 1, 45kDa |
| chr15_+_74610894 | 0.30 |
ENST00000558821.1
ENST00000268082.4 |
CCDC33
|
coiled-coil domain containing 33 |
| chr3_-_69101413 | 0.28 |
ENST00000398559.2
|
TMF1
|
TATA element modulatory factor 1 |
| chrX_-_135338503 | 0.27 |
ENST00000370663.5
|
MAP7D3
|
MAP7 domain containing 3 |
| chr5_-_58295712 | 0.26 |
ENST00000317118.8
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr6_+_150690028 | 0.26 |
ENST00000229447.5
ENST00000344419.3 |
IYD
|
iodotyrosine deiodinase |
| chr1_+_235530675 | 0.25 |
ENST00000366601.3
ENST00000406207.1 ENST00000543662.1 |
TBCE
|
tubulin folding cofactor E |
| chr19_+_55235969 | 0.24 |
ENST00000402254.2
ENST00000538269.1 ENST00000541392.1 ENST00000291860.1 ENST00000396284.2 |
KIR3DL1
KIR3DL3
KIR2DL4
|
killer cell immunoglobulin-like receptor, three domains, long cytoplasmic tail, 1 killer cell immunoglobulin-like receptor, three domains, long cytoplasmic tail, 3 killer cell immunoglobulin-like receptor, two domains, long cytoplasmic tail, 4 |
| chr9_+_70971815 | 0.22 |
ENST00000396392.1
ENST00000396396.1 |
PGM5
|
phosphoglucomutase 5 |
| chr20_+_56136136 | 0.21 |
ENST00000319441.4
ENST00000543666.1 |
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chrX_-_54209640 | 0.21 |
ENST00000375180.2
ENST00000328235.4 ENST00000477084.1 |
FAM120C
|
family with sequence similarity 120C |
| chr6_+_7541808 | 0.20 |
ENST00000379802.3
|
DSP
|
desmoplakin |
| chr19_+_11200038 | 0.20 |
ENST00000558518.1
ENST00000557933.1 ENST00000455727.2 ENST00000535915.1 ENST00000545707.1 ENST00000558013.1 |
LDLR
|
low density lipoprotein receptor |
| chr9_+_88556036 | 0.19 |
ENST00000361671.5
ENST00000416045.1 |
NAA35
|
N(alpha)-acetyltransferase 35, NatC auxiliary subunit |
| chr8_-_141810634 | 0.16 |
ENST00000521986.1
ENST00000523539.1 ENST00000538769.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr16_-_18441131 | 0.14 |
ENST00000339303.5
|
NPIPA8
|
nuclear pore complex interacting protein family, member A8 |
| chr1_+_111991474 | 0.13 |
ENST00000369722.3
|
ATP5F1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit B1 |
| chr4_+_95972822 | 0.10 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr17_+_66539369 | 0.08 |
ENST00000600820.1
|
AC079210.1
|
Uncharacterized protein; cDNA FLJ45097 fis, clone BRAWH3031054 |
| chr2_-_89266286 | 0.08 |
ENST00000464162.1
|
IGKV1-6
|
immunoglobulin kappa variable 1-6 |
| chr6_+_31588478 | 0.07 |
ENST00000376007.4
ENST00000376033.2 |
PRRC2A
|
proline-rich coiled-coil 2A |
| chr15_-_75871589 | 0.06 |
ENST00000306726.2
|
PTPN9
|
protein tyrosine phosphatase, non-receptor type 9 |
| chr4_-_153303658 | 0.04 |
ENST00000296555.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr8_-_102216925 | 0.04 |
ENST00000517844.1
|
ZNF706
|
zinc finger protein 706 |
| chrX_+_95939711 | 0.03 |
ENST00000373049.4
ENST00000324765.8 |
DIAPH2
|
diaphanous-related formin 2 |
| chr1_-_153521597 | 0.02 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3 |
| chr2_-_84686552 | 0.02 |
ENST00000393868.2
|
SUCLG1
|
succinate-CoA ligase, alpha subunit |
| chr2_-_31361543 | 0.02 |
ENST00000349752.5
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr16_-_66952742 | 0.01 |
ENST00000565235.2
ENST00000568632.1 ENST00000565796.1 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr16_-_66952779 | 0.01 |
ENST00000570262.1
ENST00000394055.3 ENST00000299752.4 |
CDH16
|
cadherin 16, KSP-cadherin |
| chrX_-_153775426 | 0.01 |
ENST00000393562.2
|
G6PD
|
glucose-6-phosphate dehydrogenase |
| chr10_-_48416849 | 0.00 |
ENST00000249598.1
|
GDF2
|
growth differentiation factor 2 |
| chr6_-_87804815 | 0.00 |
ENST00000369582.2
|
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr13_-_72441315 | 0.00 |
ENST00000305425.4
ENST00000313174.7 ENST00000354591.4 |
DACH1
|
dachshund homolog 1 (Drosophila) |
Network of associatons between targets according to the STRING database.
First level regulatory network of BCL6B
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0097052 | tryptophan catabolic process to acetyl-CoA(GO:0019442) L-kynurenine metabolic process(GO:0097052) |
| 0.9 | 4.5 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.7 | 2.2 | GO:0034761 | positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.7 | 4.9 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.5 | 1.6 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.5 | 3.8 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.5 | 2.8 | GO:2001202 | negative regulation of transforming growth factor-beta secretion(GO:2001202) |
| 0.5 | 3.2 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.4 | 2.5 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.4 | 1.8 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.4 | 2.8 | GO:0015866 | ADP transport(GO:0015866) |
| 0.4 | 1.8 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.3 | 4.4 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.3 | 4.8 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.3 | 4.0 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 2.5 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.2 | 1.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.2 | 0.9 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.2 | 1.7 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.2 | 1.0 | GO:0086053 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.2 | 0.9 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.2 | 1.9 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.2 | 1.9 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 1.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 1.4 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.1 | 0.6 | GO:0035936 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.1 | 0.7 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 | 2.0 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.1 | 0.6 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 1.9 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 0.3 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.1 | 0.8 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 2.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 2.3 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.1 | 0.3 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 1.0 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.1 | 0.6 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.2 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 1.8 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 0.7 | GO:0015846 | polyamine transport(GO:0015846) |
| 0.1 | 0.2 | GO:1903978 | regulation of microglial cell activation(GO:1903978) positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.1 | 1.2 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.1 | 1.4 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.1 | 2.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.2 | GO:0006114 | glycerol biosynthetic process(GO:0006114) response to methionine(GO:1904640) |
| 0.0 | 0.8 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 4.3 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.9 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.6 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.3 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 2.4 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.5 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.1 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.9 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.2 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 3.6 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.2 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 3.1 | GO:0060271 | cilium morphogenesis(GO:0060271) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.2 | 4.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 3.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 4.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 1.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.9 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 1.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 5.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.7 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 1.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 4.3 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 1.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.8 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 3.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 4.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.6 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 7.5 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 1.3 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 1.6 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 2.1 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 3.6 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.2 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 1.9 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 2.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.6 | GO:0005681 | spliceosomal complex(GO:0005681) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.8 | 3.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.7 | 3.0 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.6 | 2.5 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.5 | 2.5 | GO:0070728 | leucine binding(GO:0070728) |
| 0.5 | 1.4 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.4 | 4.5 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.4 | 1.6 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 0.4 | 4.0 | GO:0046790 | virion binding(GO:0046790) |
| 0.4 | 2.8 | GO:0000295 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) ATP transmembrane transporter activity(GO:0005347) purine nucleotide transmembrane transporter activity(GO:0015216) ADP transmembrane transporter activity(GO:0015217) |
| 0.3 | 1.9 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.3 | 0.9 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.3 | 3.6 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.3 | 2.0 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.2 | 0.7 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.2 | 1.0 | GO:0086077 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.2 | 1.3 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.2 | 0.5 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.2 | 1.0 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.2 | 0.7 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.2 | 1.6 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 2.6 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 0.2 | GO:0030109 | HLA-B specific inhibitory MHC class I receptor activity(GO:0030109) |
| 0.1 | 4.9 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 1.8 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 3.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.4 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 1.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 1.1 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 0.2 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.1 | 1.7 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.6 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 0.9 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 2.2 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.8 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.8 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 2.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 1.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 5.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 4.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 4.4 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.6 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.3 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 9.0 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 4.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 3.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 4.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 3.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 5.6 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 1.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 1.6 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.9 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.8 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 4.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 3.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 2.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.2 | 6.2 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.2 | 3.0 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 2.0 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 6.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 3.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 3.2 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 1.0 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.0 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.7 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 4.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 2.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.1 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.2 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 1.3 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.4 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |