Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
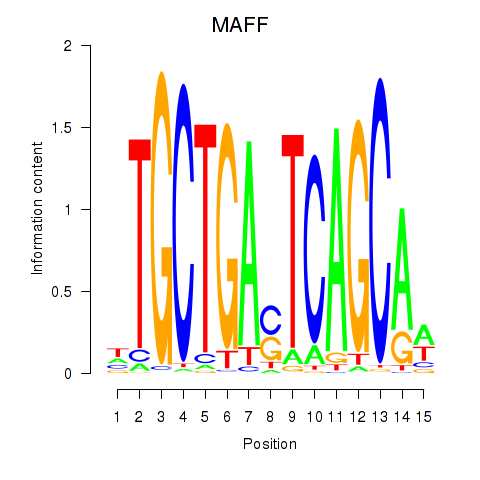
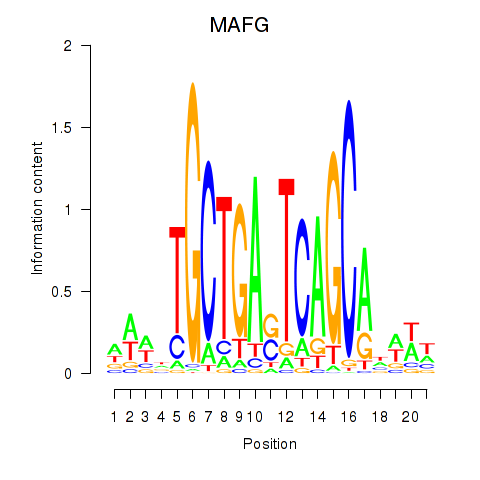
Results for MAFF_MAFG
Z-value: 0.60
Transcription factors associated with MAFF_MAFG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MAFF
|
ENSG00000185022.7 | MAF bZIP transcription factor F |
|
MAFG
|
ENSG00000197063.6 | MAF bZIP transcription factor G |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MAFG | hg19_v2_chr17_-_79885576_79885624, hg19_v2_chr17_-_79881408_79881423 | -0.10 | 1.4e-01 | Click! |
| MAFF | hg19_v2_chr22_+_38609538_38609547 | -0.02 | 7.2e-01 | Click! |
Activity profile of MAFF_MAFG motif
Sorted Z-values of MAFF_MAFG motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MAFF_MAFG
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.3 | 21.2 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 4.7 | 14.1 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 2.3 | 14.1 | GO:0070458 | cellular detoxification of nitrogen compound(GO:0070458) |
| 1.8 | 5.5 | GO:0060437 | lung growth(GO:0060437) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 1.6 | 4.9 | GO:0052331 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 1.4 | 28.9 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 1.4 | 4.2 | GO:0033214 | iron assimilation(GO:0033212) iron assimilation by chelation and transport(GO:0033214) positive regulation of bone mineralization involved in bone maturation(GO:1900159) negative regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000308) |
| 1.4 | 8.3 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 1.2 | 4.9 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 1.2 | 7.3 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 1.1 | 13.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 1.1 | 4.4 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 1.1 | 3.2 | GO:0018307 | enzyme active site formation(GO:0018307) tRNA thio-modification(GO:0034227) |
| 1.0 | 2.0 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.9 | 4.7 | GO:0090467 | L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 0.9 | 7.0 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.8 | 3.3 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.8 | 5.5 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.7 | 5.0 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.7 | 2.8 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.7 | 2.1 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.7 | 4.8 | GO:0036018 | cellular response to erythropoietin(GO:0036018) |
| 0.7 | 2.0 | GO:0097056 | seryl-tRNA aminoacylation(GO:0006434) selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.7 | 2.7 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.7 | 2.7 | GO:0032053 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.7 | 9.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.6 | 4.5 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.6 | 1.9 | GO:2000048 | positive regulation of fibrinolysis(GO:0051919) interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.6 | 8.6 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.6 | 1.2 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.6 | 1.7 | GO:0070541 | response to platinum ion(GO:0070541) |
| 0.6 | 3.4 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.6 | 3.3 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.6 | 3.3 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.5 | 3.4 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.5 | 3.9 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.5 | 0.9 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.5 | 2.3 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.5 | 3.2 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.4 | 2.2 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.4 | 5.3 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.4 | 7.2 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.4 | 2.4 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.4 | 1.2 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.4 | 4.4 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.4 | 1.2 | GO:1900158 | negative regulation of osteoclast proliferation(GO:0090291) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.4 | 3.8 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.4 | 1.5 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.4 | 1.9 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.4 | 1.5 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.4 | 1.8 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.4 | 3.3 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.3 | 1.4 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.3 | 0.7 | GO:1900110 | negative regulation of histone H3-K9 dimethylation(GO:1900110) |
| 0.3 | 1.9 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.3 | 8.9 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.3 | 1.9 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.3 | 1.3 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.3 | 5.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.3 | 2.5 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.3 | 4.6 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.3 | 2.1 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.3 | 1.7 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.3 | 5.2 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.3 | 1.4 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.3 | 1.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.3 | 0.8 | GO:0003099 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.3 | 0.8 | GO:2000282 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) regulation of cellular amino acid biosynthetic process(GO:2000282) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.3 | 1.0 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.3 | 0.8 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.3 | 34.5 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.2 | 38.9 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.2 | 1.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.2 | 2.4 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 3.5 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.2 | 1.2 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.2 | 6.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.2 | 0.7 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.2 | 1.1 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.2 | 1.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.2 | 1.5 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.2 | 2.3 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.2 | 2.7 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.2 | 1.4 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.2 | 4.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.2 | 3.8 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.2 | 2.2 | GO:2000047 | regulation of cell-cell adhesion mediated by cadherin(GO:2000047) positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.2 | 1.0 | GO:0035936 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.2 | 1.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.2 | 3.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.2 | 1.9 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.2 | 1.3 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.2 | 1.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.2 | 0.4 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.2 | 0.5 | GO:0015855 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.2 | 0.5 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.2 | 7.0 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.2 | 5.1 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.2 | 1.2 | GO:0048105 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.2 | 9.6 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.2 | 0.8 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.2 | 3.8 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.2 | 0.9 | GO:0009624 | response to nematode(GO:0009624) |
| 0.2 | 0.6 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.1 | 1.5 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 1.4 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.1 | 1.4 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.9 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 2.7 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 5.6 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.1 | 1.3 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 3.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 0.7 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 1.3 | GO:0075713 | establishment of integrated proviral latency(GO:0075713) |
| 0.1 | 4.7 | GO:0051301 | cell division(GO:0051301) |
| 0.1 | 1.2 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.8 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.1 | 1.0 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 5.5 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.1 | 9.7 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.1 | 18.1 | GO:0071346 | cellular response to interferon-gamma(GO:0071346) |
| 0.1 | 0.9 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 0.8 | GO:0035372 | protein localization to microtubule(GO:0035372) regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 1.6 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 0.6 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.1 | 6.6 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 0.4 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 0.9 | GO:0006090 | pyruvate metabolic process(GO:0006090) |
| 0.1 | 0.6 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.1 | 0.6 | GO:0051344 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.1 | 0.5 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 3.9 | GO:0007186 | G-protein coupled receptor signaling pathway(GO:0007186) |
| 0.1 | 5.0 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.1 | 0.3 | GO:0019474 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.1 | 0.9 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.1 | 6.0 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 2.6 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.1 | 2.0 | GO:0098655 | cation transmembrane transport(GO:0098655) |
| 0.1 | 0.4 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 0.9 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 1.3 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.1 | 2.8 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 6.0 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 0.7 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 0.4 | GO:0036100 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.1 | 1.5 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 0.5 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.1 | 1.2 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.1 | 2.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 1.4 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.1 | 0.5 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 4.0 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.1 | 1.7 | GO:0009081 | branched-chain amino acid metabolic process(GO:0009081) branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 2.0 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.1 | 3.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 14.0 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 0.4 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.1 | 3.4 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.8 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.0 | 2.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 1.6 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.7 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) sodium ion export from cell(GO:0036376) potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 1.2 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.5 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 1.4 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 1.0 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 1.5 | GO:0031640 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.0 | 0.3 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.9 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.5 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.3 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 2.2 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 0.2 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 3.2 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 3.4 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.0 | 2.7 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 1.8 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 0.2 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 1.5 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.0 | 5.1 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.2 | GO:0048003 | antigen processing and presentation via MHC class Ib(GO:0002475) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.4 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.0 | 0.3 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.0 | 0.9 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.0 | 0.8 | GO:0060135 | maternal process involved in female pregnancy(GO:0060135) |
| 0.0 | 1.1 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.5 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 1.1 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 1.0 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 1.3 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.0 | 0.1 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.2 | GO:0006641 | triglyceride metabolic process(GO:0006641) |
| 0.0 | 0.1 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.2 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.0 | 0.0 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 29.1 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 5.3 | 21.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 2.4 | 28.9 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 1.8 | 7.3 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 1.1 | 5.5 | GO:0001652 | granular component(GO:0001652) |
| 1.0 | 8.3 | GO:0045179 | apical cortex(GO:0045179) |
| 1.0 | 5.1 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.8 | 53.9 | GO:0005844 | polysome(GO:0005844) |
| 0.7 | 9.8 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.7 | 6.1 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.7 | 2.7 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.7 | 3.3 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.6 | 1.9 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.6 | 2.8 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.6 | 3.9 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.5 | 5.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.5 | 2.4 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.4 | 2.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.4 | 1.2 | GO:0060187 | cell pole(GO:0060187) |
| 0.4 | 8.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.4 | 2.6 | GO:0072487 | MSL complex(GO:0072487) |
| 0.4 | 4.6 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.3 | 0.7 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.3 | 3.2 | GO:0097433 | dense body(GO:0097433) |
| 0.3 | 11.8 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.2 | 3.7 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 1.5 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.2 | 1.3 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.2 | 2.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 4.5 | GO:0042627 | chylomicron(GO:0042627) |
| 0.2 | 5.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 1.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 0.6 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.1 | 2.5 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 1.9 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 1.4 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 4.1 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 4.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 3.5 | GO:0036019 | endolysosome(GO:0036019) |
| 0.1 | 1.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 2.6 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 2.6 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.1 | 2.2 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 7.2 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 1.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 5.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 2.8 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.1 | 0.8 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 1.4 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 0.8 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.3 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.1 | 6.1 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 2.2 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 10.5 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 2.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 0.7 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.1 | 17.3 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 5.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 1.1 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 2.6 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 2.4 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 1.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.2 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 0.8 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 1.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 4.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 5.6 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 3.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 0.7 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.1 | 6.1 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 1.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 16.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 16.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 4.4 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.1 | 5.3 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 6.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 3.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 1.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.5 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 7.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 1.6 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 2.0 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 4.8 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 29.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 4.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 1.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 3.7 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 1.2 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 3.3 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 3.4 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 1.5 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 4.0 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.9 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.5 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) microbody part(GO:0044438) peroxisomal part(GO:0044439) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.3 | 50.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 2.4 | 7.2 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 1.8 | 5.5 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 1.5 | 20.1 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 1.5 | 4.5 | GO:0017129 | triglyceride binding(GO:0017129) |
| 1.1 | 3.4 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 1.1 | 3.4 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 1.1 | 3.3 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.9 | 3.4 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 0.8 | 3.3 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.8 | 14.1 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.8 | 3.1 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.8 | 3.9 | GO:0070728 | leucine binding(GO:0070728) |
| 0.7 | 9.7 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.7 | 2.0 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.7 | 9.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.6 | 46.8 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.6 | 3.2 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.6 | 2.4 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.6 | 24.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.5 | 2.2 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.5 | 5.3 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.5 | 1.9 | GO:0050473 | arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.5 | 1.8 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.5 | 8.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.4 | 1.3 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.4 | 5.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.4 | 10.9 | GO:0005521 | lamin binding(GO:0005521) |
| 0.4 | 2.1 | GO:0070404 | NADH binding(GO:0070404) |
| 0.4 | 5.8 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.4 | 2.0 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.4 | 4.4 | GO:0004568 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.4 | 5.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.4 | 1.5 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.4 | 3.0 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.3 | 9.4 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.3 | 4.7 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.3 | 4.9 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.3 | 3.6 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.3 | 2.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.3 | 6.1 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.3 | 0.9 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.3 | 0.8 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.3 | 3.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.3 | 8.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.3 | 10.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.3 | 0.8 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.3 | 2.9 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.2 | 1.5 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.2 | 1.9 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.2 | 8.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 0.7 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.2 | 2.6 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.2 | 6.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 7.1 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.2 | 1.0 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.2 | 1.8 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.2 | 4.9 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 1.9 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.2 | 5.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.2 | 1.5 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.2 | 2.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 0.5 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.2 | 0.5 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.2 | 1.2 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.2 | 0.5 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.2 | 1.2 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.2 | 15.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 2.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 42.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.0 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 6.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.4 | GO:0097259 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.1 | 0.9 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 1.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 4.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 1.6 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 2.9 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 2.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 1.8 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 2.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 8.8 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 1.2 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.7 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.1 | 7.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 0.6 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.7 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.6 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 4.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 2.6 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 0.3 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 1.0 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 0.8 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.1 | 1.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 0.5 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 2.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 1.2 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.1 | 1.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 2.1 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 0.8 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.1 | 0.7 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 2.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 1.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 1.1 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 2.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 7.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 5.2 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 2.0 | GO:0051119 | sugar transmembrane transporter activity(GO:0051119) |
| 0.0 | 1.5 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.3 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 1.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.9 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.6 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 2.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.7 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 3.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 1.5 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 2.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 1.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 2.1 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.7 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.2 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 1.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0030883 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.4 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.5 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.9 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.8 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 1.3 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.5 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 41.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 13.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 7.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 5.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 5.5 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.2 | 5.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.2 | 18.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 6.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 0.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 5.6 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 4.4 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 4.7 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 5.3 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 2.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 1.9 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 1.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 15.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 2.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.9 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.8 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 2.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 2.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.3 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 2.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 2.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 6.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 4.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 2.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.9 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 29.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.8 | 9.7 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.7 | 11.0 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.6 | 44.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.5 | 2.0 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.5 | 6.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.4 | 8.9 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.4 | 9.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.4 | 8.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.4 | 20.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.4 | 8.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.3 | 11.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.3 | 6.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 3.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.3 | 2.8 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.3 | 2.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 3.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 14.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 3.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.2 | 5.1 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.2 | 2.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 2.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 13.0 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.2 | 2.7 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 15.7 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.1 | 3.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 11.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 2.0 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 1.5 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.1 | 4.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.9 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.1 | 5.6 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 9.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 1.3 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 1.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 3.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 0.7 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 2.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 2.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 2.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.9 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.8 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 1.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.9 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 3.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.9 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.5 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 4.2 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 2.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |