Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
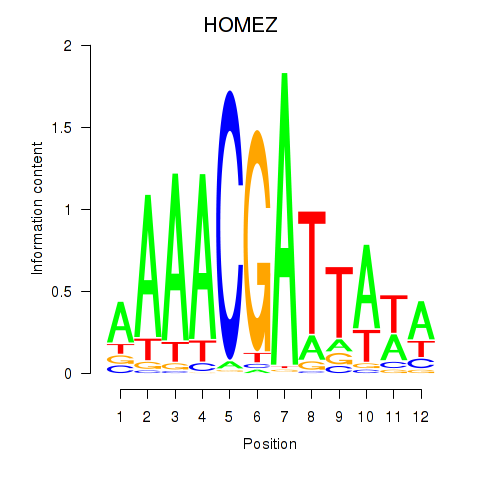
Results for HOMEZ
Z-value: 1.13
Transcription factors associated with HOMEZ
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOMEZ
|
ENSG00000215271.6 | homeobox and leucine zipper encoding |
Activity profile of HOMEZ motif
Sorted Z-values of HOMEZ motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_95055956 | 35.43 |
ENST00000375629.3
ENST00000447699.2 ENST00000375643.3 ENST00000395554.3 |
IARS
|
isoleucyl-tRNA synthetase |
| chr7_-_7679633 | 31.08 |
ENST00000401447.1
|
RPA3
|
replication protein A3, 14kDa |
| chr3_+_33155525 | 28.37 |
ENST00000449224.1
|
CRTAP
|
cartilage associated protein |
| chr10_-_27443294 | 28.33 |
ENST00000396296.3
ENST00000375972.3 ENST00000376016.3 ENST00000491542.2 |
YME1L1
|
YME1-like 1 ATPase |
| chr9_-_95056010 | 24.84 |
ENST00000443024.2
|
IARS
|
isoleucyl-tRNA synthetase |
| chr3_-_64009658 | 24.23 |
ENST00000394431.2
|
PSMD6
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chrX_-_135962876 | 22.83 |
ENST00000431446.3
ENST00000570135.1 ENST00000320676.7 ENST00000562646.1 |
RBMX
|
RNA binding motif protein, X-linked |
| chr11_+_102217936 | 21.71 |
ENST00000532832.1
ENST00000530675.1 ENST00000533742.1 ENST00000227758.2 ENST00000532672.1 ENST00000531259.1 ENST00000527465.1 |
BIRC2
|
baculoviral IAP repeat containing 2 |
| chr2_+_187371440 | 21.54 |
ENST00000445547.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr8_-_54935001 | 21.02 |
ENST00000396401.3
ENST00000521604.2 |
TCEA1
|
transcription elongation factor A (SII), 1 |
| chr20_-_5107180 | 20.42 |
ENST00000379160.3
|
PCNA
|
proliferating cell nuclear antigen |
| chr11_-_62607036 | 20.18 |
ENST00000311713.7
ENST00000278856.4 |
WDR74
|
WD repeat domain 74 |
| chr5_+_72143988 | 19.27 |
ENST00000506351.2
|
TNPO1
|
transportin 1 |
| chr4_-_103747011 | 19.11 |
ENST00000350435.7
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr1_+_113161778 | 18.96 |
ENST00000263168.3
|
CAPZA1
|
capping protein (actin filament) muscle Z-line, alpha 1 |
| chrX_+_119737806 | 18.71 |
ENST00000371317.5
|
MCTS1
|
malignant T cell amplified sequence 1 |
| chr13_-_31038370 | 18.47 |
ENST00000399489.1
ENST00000339872.4 |
HMGB1
|
high mobility group box 1 |
| chr1_+_24018269 | 17.79 |
ENST00000374550.3
|
RPL11
|
ribosomal protein L11 |
| chr4_-_103746683 | 17.70 |
ENST00000504211.1
ENST00000508476.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr4_-_174254823 | 17.57 |
ENST00000438704.2
|
HMGB2
|
high mobility group box 2 |
| chr6_+_34725263 | 17.10 |
ENST00000374018.1
ENST00000374017.3 |
SNRPC
|
small nuclear ribonucleoprotein polypeptide C |
| chr14_+_58711539 | 17.00 |
ENST00000216455.4
ENST00000412908.2 ENST00000557508.1 |
PSMA3
|
proteasome (prosome, macropain) subunit, alpha type, 3 |
| chr20_+_30327063 | 16.99 |
ENST00000300403.6
ENST00000340513.4 |
TPX2
|
TPX2, microtubule-associated |
| chr8_-_101962777 | 16.85 |
ENST00000395951.3
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr4_-_103746924 | 16.51 |
ENST00000505207.1
ENST00000502404.1 ENST00000507845.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr2_+_242289502 | 16.39 |
ENST00000451310.1
|
SEPT2
|
septin 2 |
| chr1_-_165738072 | 15.44 |
ENST00000481278.1
|
TMCO1
|
transmembrane and coiled-coil domains 1 |
| chr3_-_146262488 | 15.41 |
ENST00000487389.1
|
PLSCR1
|
phospholipid scramblase 1 |
| chr7_+_98923505 | 15.29 |
ENST00000432884.2
ENST00000262942.5 |
ARPC1A
|
actin related protein 2/3 complex, subunit 1A, 41kDa |
| chr10_-_72648541 | 14.91 |
ENST00000299299.3
|
PCBD1
|
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha |
| chr2_-_150444116 | 14.90 |
ENST00000428879.1
ENST00000422782.2 |
MMADHC
|
methylmalonic aciduria (cobalamin deficiency) cblD type, with homocystinuria |
| chr10_-_121296045 | 14.72 |
ENST00000392865.1
|
RGS10
|
regulator of G-protein signaling 10 |
| chr14_-_105420241 | 14.34 |
ENST00000557457.1
|
AHNAK2
|
AHNAK nucleoprotein 2 |
| chr4_+_140222609 | 14.30 |
ENST00000296543.5
ENST00000398947.1 |
NAA15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr2_+_233415363 | 14.23 |
ENST00000409514.1
ENST00000409098.1 ENST00000409495.1 ENST00000409167.3 ENST00000409322.1 ENST00000409394.1 |
EIF4E2
|
eukaryotic translation initiation factor 4E family member 2 |
| chr1_+_220267429 | 14.19 |
ENST00000366922.1
ENST00000302637.5 |
IARS2
|
isoleucyl-tRNA synthetase 2, mitochondrial |
| chr2_-_225362533 | 13.87 |
ENST00000451538.1
|
CUL3
|
cullin 3 |
| chr1_-_54405773 | 13.79 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr1_-_19426149 | 13.77 |
ENST00000429347.2
|
UBR4
|
ubiquitin protein ligase E3 component n-recognin 4 |
| chr6_+_34725181 | 13.77 |
ENST00000244520.5
|
SNRPC
|
small nuclear ribonucleoprotein polypeptide C |
| chr10_-_95242044 | 13.68 |
ENST00000371501.4
ENST00000371502.4 ENST00000371489.1 |
MYOF
|
myoferlin |
| chr16_-_23607598 | 13.63 |
ENST00000562133.1
ENST00000570319.1 ENST00000007516.3 |
NDUFAB1
|
NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1, 8kDa |
| chr20_+_44441626 | 13.37 |
ENST00000372568.4
|
UBE2C
|
ubiquitin-conjugating enzyme E2C |
| chrX_+_12993336 | 13.32 |
ENST00000380635.1
|
TMSB4X
|
thymosin beta 4, X-linked |
| chr2_+_109237717 | 13.29 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chrX_-_119709637 | 13.28 |
ENST00000404115.3
|
CUL4B
|
cullin 4B |
| chr1_-_193075180 | 13.22 |
ENST00000367440.3
|
GLRX2
|
glutaredoxin 2 |
| chr4_-_104119528 | 13.20 |
ENST00000380026.3
ENST00000503705.1 ENST00000265148.3 |
CENPE
|
centromere protein E, 312kDa |
| chr3_-_107941230 | 13.14 |
ENST00000264538.3
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr10_-_95241951 | 12.91 |
ENST00000358334.5
ENST00000359263.4 ENST00000371488.3 |
MYOF
|
myoferlin |
| chr2_+_216974020 | 12.89 |
ENST00000392132.2
ENST00000417391.1 |
XRCC5
|
X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining) |
| chr20_-_48330377 | 12.76 |
ENST00000371711.4
|
B4GALT5
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
| chr3_-_146262637 | 12.24 |
ENST00000472349.1
ENST00000342435.4 |
PLSCR1
|
phospholipid scramblase 1 |
| chr1_-_245027833 | 12.20 |
ENST00000444376.2
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) |
| chr17_-_80017856 | 12.13 |
ENST00000577574.1
|
DUS1L
|
dihydrouridine synthase 1-like (S. cerevisiae) |
| chr11_+_101983176 | 12.08 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chr17_-_8113542 | 12.08 |
ENST00000578549.1
ENST00000535053.1 ENST00000582368.1 |
AURKB
|
aurora kinase B |
| chr12_-_50419177 | 12.01 |
ENST00000454520.2
ENST00000546595.1 ENST00000548824.1 ENST00000549777.1 ENST00000546723.1 ENST00000427314.2 ENST00000552157.1 ENST00000552310.1 ENST00000548644.1 ENST00000312377.5 ENST00000546786.1 ENST00000550149.1 ENST00000546764.1 ENST00000552004.1 ENST00000548320.1 ENST00000547905.1 ENST00000550651.1 ENST00000551145.1 ENST00000434422.1 ENST00000552921.1 |
RACGAP1
|
Rac GTPase activating protein 1 |
| chr19_-_49121054 | 11.86 |
ENST00000546623.1
ENST00000084795.5 |
RPL18
|
ribosomal protein L18 |
| chr4_+_83956237 | 11.86 |
ENST00000264389.2
|
COPS4
|
COP9 signalosome subunit 4 |
| chr5_+_177019159 | 11.85 |
ENST00000332598.6
|
TMED9
|
transmembrane emp24 protein transport domain containing 9 |
| chrX_+_118708493 | 11.83 |
ENST00000371558.2
|
UBE2A
|
ubiquitin-conjugating enzyme E2A |
| chr1_+_29063119 | 11.75 |
ENST00000474884.1
ENST00000542507.1 |
YTHDF2
|
YTH domain family, member 2 |
| chr7_-_7680601 | 11.71 |
ENST00000396682.2
|
RPA3
|
replication protein A3, 14kDa |
| chr4_+_83956312 | 11.66 |
ENST00000509317.1
ENST00000503682.1 ENST00000511653.1 |
COPS4
|
COP9 signalosome subunit 4 |
| chr5_+_96038476 | 11.64 |
ENST00000511049.1
ENST00000309190.5 ENST00000510156.1 ENST00000509903.1 ENST00000511782.1 ENST00000504465.1 |
CAST
|
calpastatin |
| chr2_+_118572226 | 11.61 |
ENST00000263239.2
|
DDX18
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18 |
| chr22_-_36924944 | 11.27 |
ENST00000405442.1
ENST00000402116.1 |
EIF3D
|
eukaryotic translation initiation factor 3, subunit D |
| chr7_-_102985288 | 11.19 |
ENST00000379263.3
|
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr18_-_54305658 | 11.16 |
ENST00000586262.1
ENST00000217515.6 |
TXNL1
|
thioredoxin-like 1 |
| chr1_+_144220127 | 11.06 |
ENST00000369373.5
|
NBPF8
|
neuroblastoma breakpoint family, member 8 |
| chr4_-_170679024 | 10.86 |
ENST00000393381.2
|
C4orf27
|
chromosome 4 open reading frame 27 |
| chrX_-_109590174 | 10.85 |
ENST00000372054.1
|
GNG5P2
|
guanine nucleotide binding protein (G protein), gamma 5 pseudogene 2 |
| chrX_-_114953669 | 10.78 |
ENST00000449327.1
|
RP1-241P17.4
|
Uncharacterized protein |
| chrX_-_77225135 | 10.74 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr9_-_127952032 | 10.71 |
ENST00000456642.1
ENST00000373546.3 ENST00000373547.4 |
PPP6C
|
protein phosphatase 6, catalytic subunit |
| chr15_+_65843130 | 10.64 |
ENST00000569894.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr1_-_245027766 | 10.55 |
ENST00000283179.9
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) |
| chrX_+_118708517 | 10.54 |
ENST00000346330.3
|
UBE2A
|
ubiquitin-conjugating enzyme E2A |
| chr16_-_58585513 | 10.47 |
ENST00000245138.4
ENST00000567285.1 |
CNOT1
|
CCR4-NOT transcription complex, subunit 1 |
| chr11_-_47664072 | 10.32 |
ENST00000542981.1
ENST00000530428.1 ENST00000302503.3 |
MTCH2
|
mitochondrial carrier 2 |
| chr12_+_72148614 | 10.12 |
ENST00000261263.3
|
RAB21
|
RAB21, member RAS oncogene family |
| chr10_+_85899196 | 10.06 |
ENST00000372134.3
|
GHITM
|
growth hormone inducible transmembrane protein |
| chr17_+_66521936 | 10.03 |
ENST00000592800.1
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chrX_-_151999269 | 9.94 |
ENST00000370277.3
|
CETN2
|
centrin, EF-hand protein, 2 |
| chr6_+_44214824 | 9.78 |
ENST00000371646.5
ENST00000353801.3 |
HSP90AB1
|
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chr4_+_113568207 | 9.61 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr3_-_64009102 | 9.56 |
ENST00000478185.1
ENST00000482510.1 ENST00000497323.1 ENST00000492933.1 ENST00000295901.4 |
PSMD6
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chr2_+_85843252 | 9.52 |
ENST00000409025.1
ENST00000409470.1 ENST00000323701.6 ENST00000409766.3 |
USP39
|
ubiquitin specific peptidase 39 |
| chr16_+_74330673 | 9.48 |
ENST00000219313.4
ENST00000540379.1 ENST00000567958.1 ENST00000568615.2 |
PSMD7
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 7 |
| chr5_+_112196919 | 9.46 |
ENST00000505459.1
ENST00000282999.3 ENST00000515463.1 |
SRP19
|
signal recognition particle 19kDa |
| chr18_-_47018897 | 9.45 |
ENST00000418495.1
|
RPL17
|
ribosomal protein L17 |
| chr2_+_47630108 | 9.40 |
ENST00000233146.2
ENST00000454849.1 ENST00000543555.1 |
MSH2
|
mutS homolog 2 |
| chr18_+_3247413 | 9.37 |
ENST00000579226.1
ENST00000217652.3 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr10_-_123734683 | 9.25 |
ENST00000369017.5
ENST00000369023.3 |
NSMCE4A
|
non-SMC element 4 homolog A (S. cerevisiae) |
| chr18_+_20513278 | 9.23 |
ENST00000327155.5
|
RBBP8
|
retinoblastoma binding protein 8 |
| chr8_+_91013577 | 9.17 |
ENST00000220764.2
|
DECR1
|
2,4-dienoyl CoA reductase 1, mitochondrial |
| chr9_-_128412696 | 9.08 |
ENST00000420643.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr1_-_68962782 | 9.08 |
ENST00000456315.2
|
DEPDC1
|
DEP domain containing 1 |
| chr2_-_55237484 | 8.93 |
ENST00000394609.2
|
RTN4
|
reticulon 4 |
| chr22_+_42017987 | 8.91 |
ENST00000405506.1
|
XRCC6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr3_-_10362725 | 8.89 |
ENST00000397109.3
ENST00000428626.1 ENST00000445064.1 ENST00000431352.1 ENST00000397117.1 ENST00000337354.4 ENST00000383801.2 ENST00000432213.1 ENST00000350697.3 |
SEC13
|
SEC13 homolog (S. cerevisiae) |
| chr18_-_47017956 | 8.78 |
ENST00000584895.1
ENST00000332968.6 ENST00000580210.1 ENST00000579408.1 |
RPL17-C18orf32
RPL17
|
RPL17-C18orf32 readthrough ribosomal protein L17 |
| chr7_-_102985035 | 8.71 |
ENST00000426036.2
ENST00000249270.7 ENST00000454277.1 ENST00000412522.1 |
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr1_-_153958805 | 8.53 |
ENST00000368575.3
|
RAB13
|
RAB13, member RAS oncogene family |
| chr5_+_95066823 | 8.46 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr6_+_138725343 | 8.43 |
ENST00000607197.1
ENST00000367697.3 |
HEBP2
|
heme binding protein 2 |
| chr5_-_96518907 | 8.43 |
ENST00000508447.1
ENST00000283109.3 |
RIOK2
|
RIO kinase 2 |
| chr5_-_52405564 | 8.39 |
ENST00000510818.2
ENST00000396954.3 ENST00000508922.1 ENST00000361377.4 ENST00000582677.1 ENST00000584946.1 ENST00000450852.3 |
MOCS2
|
molybdenum cofactor synthesis 2 |
| chr3_-_146262352 | 8.36 |
ENST00000462666.1
|
PLSCR1
|
phospholipid scramblase 1 |
| chr16_-_69373396 | 8.24 |
ENST00000562595.1
ENST00000562081.1 ENST00000306875.4 |
COG8
|
component of oligomeric golgi complex 8 |
| chr4_+_57845024 | 8.21 |
ENST00000431623.2
ENST00000441246.2 |
POLR2B
|
polymerase (RNA) II (DNA directed) polypeptide B, 140kDa |
| chr1_-_153950164 | 8.13 |
ENST00000271843.4
|
JTB
|
jumping translocation breakpoint |
| chr1_-_153518270 | 8.10 |
ENST00000354332.4
ENST00000368716.4 |
S100A4
|
S100 calcium binding protein A4 |
| chr4_-_106629796 | 8.05 |
ENST00000416543.1
ENST00000515819.1 ENST00000420368.2 ENST00000503746.1 ENST00000340139.5 ENST00000433009.1 |
INTS12
|
integrator complex subunit 12 |
| chr9_+_106856831 | 8.01 |
ENST00000303219.8
ENST00000374787.3 |
SMC2
|
structural maintenance of chromosomes 2 |
| chr2_-_178129853 | 7.97 |
ENST00000397062.3
|
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr15_+_77223960 | 7.93 |
ENST00000394885.3
|
RCN2
|
reticulocalbin 2, EF-hand calcium binding domain |
| chr1_-_153949751 | 7.84 |
ENST00000428469.1
|
JTB
|
jumping translocation breakpoint |
| chr1_-_153950098 | 7.77 |
ENST00000356648.1
|
JTB
|
jumping translocation breakpoint |
| chr1_-_40349106 | 7.75 |
ENST00000545233.1
ENST00000537440.1 ENST00000537223.1 ENST00000541099.1 ENST00000441669.2 ENST00000544981.1 ENST00000316891.5 ENST00000372818.1 |
TRIT1
|
tRNA isopentenyltransferase 1 |
| chr1_+_212208919 | 7.74 |
ENST00000366991.4
ENST00000542077.1 |
DTL
|
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr7_+_100303676 | 7.68 |
ENST00000303151.4
|
POP7
|
processing of precursor 7, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr9_-_128003606 | 7.64 |
ENST00000324460.6
|
HSPA5
|
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) |
| chr6_+_64281906 | 7.63 |
ENST00000370651.3
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr1_-_153950116 | 7.56 |
ENST00000368589.1
|
JTB
|
jumping translocation breakpoint |
| chr22_-_32058166 | 7.49 |
ENST00000435900.1
ENST00000336566.4 |
PISD
|
phosphatidylserine decarboxylase |
| chr6_-_33282163 | 7.49 |
ENST00000434618.2
ENST00000456592.2 |
TAPBP
|
TAP binding protein (tapasin) |
| chr22_-_32808194 | 7.43 |
ENST00000451746.2
ENST00000216038.5 |
RTCB
|
RNA 2',3'-cyclic phosphate and 5'-OH ligase |
| chr15_+_89787180 | 7.41 |
ENST00000300027.8
ENST00000310775.7 ENST00000567891.1 ENST00000564920.1 ENST00000565255.1 ENST00000567996.1 ENST00000451393.2 ENST00000563250.1 |
FANCI
|
Fanconi anemia, complementation group I |
| chr8_+_132916318 | 7.39 |
ENST00000254624.5
ENST00000522709.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chr8_-_13134045 | 7.33 |
ENST00000512044.2
|
DLC1
|
deleted in liver cancer 1 |
| chr6_-_43027105 | 7.23 |
ENST00000230413.5
ENST00000487429.1 ENST00000489623.1 ENST00000468957.1 |
MRPL2
|
mitochondrial ribosomal protein L2 |
| chr12_+_19282643 | 7.20 |
ENST00000317589.4
ENST00000355397.3 ENST00000359180.3 ENST00000309364.4 ENST00000540972.1 ENST00000429027.2 |
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr17_+_30469473 | 7.18 |
ENST00000333942.6
ENST00000358365.3 ENST00000583994.1 ENST00000545287.2 |
RHOT1
|
ras homolog family member T1 |
| chr1_-_6420737 | 7.15 |
ENST00000541130.1
ENST00000377845.3 |
ACOT7
|
acyl-CoA thioesterase 7 |
| chr12_-_65153175 | 7.07 |
ENST00000543646.1
ENST00000542058.1 ENST00000258145.3 |
GNS
|
glucosamine (N-acetyl)-6-sulfatase |
| chr1_-_86174065 | 7.00 |
ENST00000370574.3
ENST00000431532.2 |
ZNHIT6
|
zinc finger, HIT-type containing 6 |
| chr6_-_109702885 | 6.99 |
ENST00000504373.1
|
CD164
|
CD164 molecule, sialomucin |
| chr2_+_11674213 | 6.97 |
ENST00000381486.2
|
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr10_-_126849068 | 6.97 |
ENST00000494626.2
ENST00000337195.5 |
CTBP2
|
C-terminal binding protein 2 |
| chr22_-_22901477 | 6.95 |
ENST00000420709.1
ENST00000398741.1 ENST00000405655.3 |
PRAME
|
preferentially expressed antigen in melanoma |
| chr12_-_92539614 | 6.92 |
ENST00000256015.3
|
BTG1
|
B-cell translocation gene 1, anti-proliferative |
| chr2_-_37068530 | 6.89 |
ENST00000593798.1
|
AC007382.1
|
Uncharacterized protein |
| chr14_-_74959978 | 6.89 |
ENST00000541064.1
|
NPC2
|
Niemann-Pick disease, type C2 |
| chr5_-_140700322 | 6.83 |
ENST00000313368.5
|
TAF7
|
TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 55kDa |
| chr14_-_74960030 | 6.83 |
ENST00000553490.1
ENST00000557510.1 |
NPC2
|
Niemann-Pick disease, type C2 |
| chr10_+_51572339 | 6.78 |
ENST00000344348.6
|
NCOA4
|
nuclear receptor coactivator 4 |
| chr7_+_89783689 | 6.73 |
ENST00000297205.2
|
STEAP1
|
six transmembrane epithelial antigen of the prostate 1 |
| chr14_-_74959994 | 6.68 |
ENST00000238633.2
ENST00000434013.2 |
NPC2
|
Niemann-Pick disease, type C2 |
| chr1_+_44401479 | 6.66 |
ENST00000438616.3
|
ARTN
|
artemin |
| chr20_+_61299155 | 6.55 |
ENST00000451793.1
|
SLCO4A1
|
solute carrier organic anion transporter family, member 4A1 |
| chr22_-_22901636 | 6.50 |
ENST00000406503.1
ENST00000439106.1 ENST00000402697.1 ENST00000543184.1 ENST00000398743.2 |
PRAME
|
preferentially expressed antigen in melanoma |
| chrX_+_49160148 | 6.45 |
ENST00000407599.3
|
GAGE10
|
G antigen 10 |
| chr3_-_98312548 | 6.35 |
ENST00000264193.2
|
CPOX
|
coproporphyrinogen oxidase |
| chr14_-_45603657 | 6.34 |
ENST00000396062.3
|
FKBP3
|
FK506 binding protein 3, 25kDa |
| chr1_+_40204538 | 6.25 |
ENST00000324379.5
ENST00000356511.2 ENST00000497370.1 ENST00000470213.1 ENST00000372835.5 ENST00000372830.1 |
PPIE
|
peptidylprolyl isomerase E (cyclophilin E) |
| chr16_+_30075463 | 6.24 |
ENST00000562168.1
ENST00000569545.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr11_+_74303575 | 6.23 |
ENST00000263681.2
|
POLD3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr6_-_75960024 | 6.19 |
ENST00000370081.2
|
COX7A2
|
cytochrome c oxidase subunit VIIa polypeptide 2 (liver) |
| chr4_+_57845043 | 6.18 |
ENST00000433463.1
ENST00000314595.5 |
POLR2B
|
polymerase (RNA) II (DNA directed) polypeptide B, 140kDa |
| chr11_+_58910295 | 6.16 |
ENST00000420244.1
|
FAM111A
|
family with sequence similarity 111, member A |
| chr8_-_62602327 | 6.13 |
ENST00000445642.3
ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH
|
aspartate beta-hydroxylase |
| chr12_-_71551652 | 6.12 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chr7_+_117824210 | 6.11 |
ENST00000422760.1
ENST00000411938.1 |
NAA38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit |
| chr1_+_74701062 | 6.02 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr1_+_89246647 | 6.01 |
ENST00000544045.1
|
PKN2
|
protein kinase N2 |
| chr7_+_101460882 | 6.00 |
ENST00000292535.7
ENST00000549414.2 ENST00000550008.2 ENST00000546411.2 ENST00000556210.1 |
CUX1
|
cut-like homeobox 1 |
| chr3_-_133969437 | 5.96 |
ENST00000460933.1
ENST00000296084.4 |
RYK
|
receptor-like tyrosine kinase |
| chr7_+_128379346 | 5.94 |
ENST00000535011.2
ENST00000542996.2 ENST00000535623.1 ENST00000538546.1 ENST00000249364.4 ENST00000449187.2 |
CALU
|
calumenin |
| chr12_-_10978957 | 5.93 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr1_-_6259641 | 5.92 |
ENST00000234875.4
|
RPL22
|
ribosomal protein L22 |
| chr12_+_112451120 | 5.91 |
ENST00000261735.3
ENST00000455836.1 |
ERP29
|
endoplasmic reticulum protein 29 |
| chr12_-_8803128 | 5.91 |
ENST00000543467.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chr6_+_31633011 | 5.87 |
ENST00000375885.4
|
CSNK2B
|
casein kinase 2, beta polypeptide |
| chr16_-_18801643 | 5.84 |
ENST00000322989.4
ENST00000563390.1 |
RPS15A
|
ribosomal protein S15a |
| chr11_+_8008867 | 5.77 |
ENST00000309828.4
ENST00000449102.2 |
EIF3F
|
eukaryotic translation initiation factor 3, subunit F |
| chr17_+_41150290 | 5.77 |
ENST00000589037.1
ENST00000253788.5 |
RPL27
|
ribosomal protein L27 |
| chr20_+_1099233 | 5.77 |
ENST00000246015.4
ENST00000335877.6 ENST00000438768.2 |
PSMF1
|
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
| chr1_-_143767881 | 5.77 |
ENST00000419275.1
|
PPIAL4G
|
peptidylprolyl isomerase A (cyclophilin A)-like 4G |
| chr2_-_3523507 | 5.66 |
ENST00000327435.6
|
ADI1
|
acireductone dioxygenase 1 |
| chr15_+_52155001 | 5.63 |
ENST00000544199.1
|
TMOD3
|
tropomodulin 3 (ubiquitous) |
| chr10_-_27149904 | 5.57 |
ENST00000376166.1
ENST00000376138.3 ENST00000355394.4 ENST00000346832.5 ENST00000376134.3 ENST00000376137.4 ENST00000536334.1 ENST00000490841.2 |
ABI1
|
abl-interactor 1 |
| chr5_+_141488070 | 5.54 |
ENST00000253814.4
|
NDFIP1
|
Nedd4 family interacting protein 1 |
| chr10_-_126849588 | 5.49 |
ENST00000411419.2
|
CTBP2
|
C-terminal binding protein 2 |
| chr10_+_5488564 | 5.47 |
ENST00000449083.1
ENST00000380359.3 |
NET1
|
neuroepithelial cell transforming 1 |
| chr18_-_47018869 | 5.46 |
ENST00000583036.1
ENST00000580261.1 |
RPL17
|
ribosomal protein L17 |
| chr20_-_31989307 | 5.43 |
ENST00000473997.1
ENST00000544843.1 ENST00000346416.2 ENST00000357886.4 ENST00000339269.5 |
CDK5RAP1
|
CDK5 regulatory subunit associated protein 1 |
| chr16_+_88869621 | 5.41 |
ENST00000301019.4
|
CDT1
|
chromatin licensing and DNA replication factor 1 |
| chr10_+_51572408 | 5.39 |
ENST00000374082.1
|
NCOA4
|
nuclear receptor coactivator 4 |
| chr12_-_52911718 | 5.38 |
ENST00000548409.1
|
KRT5
|
keratin 5 |
| chr5_+_85913721 | 5.32 |
ENST00000247655.3
ENST00000509578.1 ENST00000515763.1 |
COX7C
|
cytochrome c oxidase subunit VIIc |
| chr22_-_31364187 | 5.32 |
ENST00000215862.4
ENST00000397641.3 |
MORC2
|
MORC family CW-type zinc finger 2 |
| chr1_-_165738085 | 5.31 |
ENST00000464650.1
ENST00000392129.6 |
TMCO1
|
transmembrane and coiled-coil domains 1 |
| chr20_-_5093713 | 5.21 |
ENST00000342308.5
ENST00000202834.7 |
TMEM230
|
transmembrane protein 230 |
| chr4_+_169753156 | 5.07 |
ENST00000393726.3
ENST00000507735.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr5_+_147774275 | 5.03 |
ENST00000513826.1
|
FBXO38
|
F-box protein 38 |
| chr12_+_100594557 | 5.00 |
ENST00000546902.1
ENST00000552376.1 ENST00000551617.1 |
ACTR6
|
ARP6 actin-related protein 6 homolog (yeast) |
| chr19_-_39330818 | 4.99 |
ENST00000594769.1
ENST00000602021.1 |
AC104534.3
|
Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial |
| chr14_+_23235886 | 4.99 |
ENST00000604262.1
ENST00000431881.2 ENST00000412791.1 ENST00000358043.5 |
OXA1L
|
oxidase (cytochrome c) assembly 1-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOMEZ
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 24.8 | 74.5 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 10.0 | 30.1 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 7.2 | 21.7 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 6.6 | 19.9 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 6.4 | 12.9 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 6.2 | 18.7 | GO:0002188 | translation reinitiation(GO:0002188) |
| 5.6 | 11.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 5.1 | 20.4 | GO:1902990 | leading strand elongation(GO:0006272) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 5.1 | 20.4 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 4.7 | 28.3 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 4.6 | 18.5 | GO:0002840 | plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) regulation of restriction endodeoxyribonuclease activity(GO:0032072) negative regulation of apoptotic cell clearance(GO:2000426) |
| 4.5 | 36.0 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 4.4 | 17.8 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 4.0 | 12.1 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 3.5 | 13.9 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 3.4 | 30.9 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 3.4 | 40.9 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 3.3 | 13.2 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 3.2 | 22.7 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 3.2 | 9.5 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 3.1 | 9.4 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 3.0 | 12.0 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 2.9 | 17.1 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 2.8 | 8.5 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 2.8 | 8.4 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 2.7 | 10.7 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 2.6 | 2.6 | GO:0006293 | nucleotide-excision repair, preincision complex stabilization(GO:0006293) nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 2.6 | 28.4 | GO:0006983 | ER overload response(GO:0006983) |
| 2.4 | 9.8 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 2.4 | 14.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 2.4 | 9.5 | GO:0061643 | chemorepulsion of dopaminergic neuron axon(GO:0036518) chemorepulsion of axon(GO:0061643) |
| 2.3 | 4.6 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 2.2 | 13.2 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 2.1 | 49.9 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
| 2.0 | 6.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 2.0 | 12.1 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 2.0 | 8.0 | GO:1901377 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 2.0 | 5.9 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 1.9 | 89.1 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 1.8 | 5.5 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 1.8 | 18.1 | GO:0042262 | DNA protection(GO:0042262) |
| 1.8 | 5.4 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 1.8 | 7.1 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 1.8 | 17.6 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 1.7 | 8.6 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 1.7 | 23.9 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 1.7 | 8.5 | GO:0030047 | actin modification(GO:0030047) |
| 1.7 | 11.7 | GO:1904688 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 1.7 | 5.0 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 1.7 | 14.9 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 1.6 | 19.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 1.6 | 12.5 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 1.5 | 9.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 1.5 | 5.9 | GO:0043335 | protein unfolding(GO:0043335) |
| 1.5 | 23.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 1.4 | 4.3 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 1.4 | 13.6 | GO:0009249 | protein lipoylation(GO:0009249) |
| 1.3 | 12.1 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 1.3 | 8.0 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 1.3 | 6.7 | GO:0097021 | Peyer's patch morphogenesis(GO:0061146) lymphocyte migration into lymphoid organs(GO:0097021) |
| 1.3 | 9.2 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 1.3 | 13.2 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 1.2 | 5.0 | GO:0031179 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 1.2 | 3.7 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 1.2 | 7.2 | GO:1902513 | regulation of organelle transport along microtubule(GO:1902513) |
| 1.2 | 2.4 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 1.2 | 26.9 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 1.2 | 7.0 | GO:0048254 | snoRNA localization(GO:0048254) |
| 1.1 | 16.8 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 1.1 | 8.9 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 1.1 | 3.3 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 1.0 | 4.2 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 1.0 | 22.8 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 1.0 | 23.2 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 1.0 | 8.0 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 1.0 | 8.9 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 1.0 | 6.7 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.9 | 5.6 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.9 | 3.4 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.8 | 4.2 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.8 | 14.7 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.8 | 3.2 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.8 | 6.9 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.7 | 4.5 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.7 | 5.9 | GO:0033211 | positive regulation of activin receptor signaling pathway(GO:0032927) adiponectin-activated signaling pathway(GO:0033211) |
| 0.7 | 41.7 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.7 | 10.0 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.7 | 4.8 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.7 | 6.8 | GO:0015846 | polyamine transport(GO:0015846) |
| 0.7 | 61.4 | GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) |
| 0.7 | 2.7 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.7 | 2.6 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.6 | 6.3 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.6 | 19.0 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.6 | 3.8 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.6 | 5.7 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.6 | 7.4 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.6 | 8.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.6 | 10.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.6 | 14.9 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.5 | 2.2 | GO:0036289 | peptidyl-serine autophosphorylation(GO:0036289) |
| 0.5 | 3.7 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.5 | 2.6 | GO:0050906 | detection of stimulus involved in sensory perception(GO:0050906) detection of chemical stimulus involved in sensory perception(GO:0050907) detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.5 | 7.2 | GO:0061458 | reproductive system development(GO:0061458) |
| 0.5 | 6.2 | GO:0045071 | regulation of viral genome replication(GO:0045069) negative regulation of viral genome replication(GO:0045071) |
| 0.5 | 7.7 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.5 | 6.5 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.5 | 2.5 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.5 | 5.8 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.5 | 4.1 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.4 | 11.6 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.4 | 8.9 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.4 | 7.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.4 | 2.2 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.4 | 23.6 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.4 | 2.5 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.4 | 8.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.4 | 61.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.4 | 3.3 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.4 | 7.5 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.4 | 5.0 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.4 | 3.7 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.4 | 4.5 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.4 | 12.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.4 | 8.1 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.4 | 1.2 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.4 | 17.0 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.4 | 18.9 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.4 | 3.0 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.4 | 2.6 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.4 | 7.4 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.4 | 13.3 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) positive regulation of adherens junction organization(GO:1903393) |
| 0.3 | 2.7 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.3 | 0.9 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.3 | 1.7 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.3 | 3.1 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.3 | 12.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.3 | 6.0 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.3 | 3.9 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.3 | 3.1 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.3 | 3.0 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.2 | 6.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 7.2 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.2 | 37.3 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.2 | 1.1 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.2 | 7.0 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.2 | 0.9 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.2 | 4.5 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.2 | 24.7 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.2 | 5.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 0.5 | GO:0042727 | flavin-containing compound biosynthetic process(GO:0042727) |
| 0.2 | 3.6 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.2 | 5.9 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.2 | 6.0 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.2 | 0.5 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.2 | 3.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.2 | 0.8 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.2 | 5.5 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.2 | 12.1 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 1.0 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 9.5 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.1 | 1.0 | GO:0009597 | detection of virus(GO:0009597) |
| 0.1 | 3.9 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 0.8 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.1 | 14.2 | GO:0006413 | translational initiation(GO:0006413) |
| 0.1 | 1.9 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.1 | 4.5 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.1 | 9.2 | GO:2001022 | positive regulation of response to DNA damage stimulus(GO:2001022) |
| 0.1 | 2.0 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 4.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 1.0 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 9.0 | GO:0006901 | vesicle coating(GO:0006901) |
| 0.1 | 1.4 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 3.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 3.5 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.1 | 9.8 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 1.2 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.1 | 5.7 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 6.5 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 2.9 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 0.5 | GO:0060356 | leucine import(GO:0060356) |
| 0.1 | 1.4 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 0.7 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 1.7 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 3.1 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.1 | 3.1 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.1 | 1.2 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.1 | 2.4 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.1 | 2.7 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 1.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.7 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 8.2 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.1 | 9.8 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.1 | 4.7 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 4.3 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.3 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 6.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 6.3 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 7.4 | GO:0090002 | establishment of protein localization to plasma membrane(GO:0090002) |
| 0.0 | 0.6 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.4 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.1 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.0 | 0.4 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.1 | GO:0009183 | purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) dGDP metabolic process(GO:0046066) |
| 0.0 | 1.9 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 3.0 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 1.2 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 0.9 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 1.5 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.0 | 3.5 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.4 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.0 | 2.8 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.0 | 1.0 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.8 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.6 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.0 | 3.9 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 2.1 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.2 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.7 | GO:0007346 | regulation of mitotic cell cycle(GO:0007346) |
| 0.0 | 0.4 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.8 | 20.4 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 4.6 | 22.8 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 4.5 | 22.4 | GO:0033503 | HULC complex(GO:0033503) |
| 4.4 | 21.8 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 3.1 | 9.4 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 2.8 | 59.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 2.7 | 19.0 | GO:0071203 | WASH complex(GO:0071203) |
| 2.7 | 10.8 | GO:0031417 | NatC complex(GO:0031417) |
| 2.6 | 21.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 2.4 | 9.8 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 2.4 | 12.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 2.4 | 12.0 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 2.4 | 30.9 | GO:0000243 | commitment complex(GO:0000243) |
| 2.3 | 9.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 2.3 | 34.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 2.3 | 22.7 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 2.0 | 14.3 | GO:0031415 | N-terminal protein acetyltransferase complex(GO:0031414) NatA complex(GO:0031415) |
| 2.0 | 21.7 | GO:0001741 | XY body(GO:0001741) |
| 1.9 | 25.3 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 1.9 | 17.0 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 1.6 | 3.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 1.6 | 6.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 1.5 | 13.9 | GO:0005827 | polar microtubule(GO:0005827) |
| 1.4 | 17.3 | GO:0044292 | dendrite terminus(GO:0044292) |
| 1.4 | 17.0 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 1.4 | 17.0 | GO:0043203 | axon hillock(GO:0043203) |
| 1.4 | 43.3 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 1.3 | 8.9 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 1.3 | 10.1 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 1.3 | 3.8 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 1.2 | 3.7 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 1.2 | 9.5 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 1.2 | 16.4 | GO:0097227 | sperm annulus(GO:0097227) |
| 1.1 | 6.7 | GO:0042825 | TAP complex(GO:0042825) |
| 1.1 | 4.5 | GO:0030891 | VCB complex(GO:0030891) |
| 1.1 | 7.7 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 1.1 | 7.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 1.0 | 10.5 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 1.0 | 6.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 1.0 | 3.0 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 1.0 | 13.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 1.0 | 12.5 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.9 | 14.2 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.9 | 8.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.9 | 7.0 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.8 | 5.9 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.8 | 6.7 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.8 | 23.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.8 | 12.2 | GO:0044754 | autolysosome(GO:0044754) |
| 0.8 | 8.0 | GO:0000796 | condensin complex(GO:0000796) |
| 0.7 | 6.8 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.7 | 9.9 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.6 | 8.9 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.6 | 31.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.6 | 11.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.6 | 67.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.6 | 9.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.6 | 22.9 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.5 | 5.9 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.5 | 10.0 | GO:0031588 | cAMP-dependent protein kinase complex(GO:0005952) nucleotide-activated protein kinase complex(GO:0031588) |
| 0.5 | 36.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.5 | 14.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.4 | 5.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.4 | 4.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 37.4 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.3 | 2.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.3 | 11.8 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.3 | 0.9 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.3 | 26.3 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.3 | 8.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.3 | 28.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.3 | 5.8 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.3 | 5.9 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.3 | 7.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.3 | 4.7 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.2 | 1.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.2 | 12.4 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 2.7 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.2 | 26.2 | GO:0005901 | caveola(GO:0005901) |
| 0.2 | 10.4 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 7.8 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.2 | 9.3 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.2 | 31.7 | GO:0030496 | midbody(GO:0030496) |
| 0.2 | 12.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.2 | 5.7 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.2 | 4.9 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.2 | 23.2 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.2 | 4.8 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.2 | 2.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.2 | 0.9 | GO:0030894 | replisome(GO:0030894) nuclear replisome(GO:0043601) |
| 0.1 | 3.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 12.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 14.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 17.2 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 15.0 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 38.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 2.5 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 20.4 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.1 | 9.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 5.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 4.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.2 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 12.3 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.1 | 14.7 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 9.0 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.1 | 3.5 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 2.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 5.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 17.0 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.1 | 18.0 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 21.0 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 2.7 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.1 | 17.9 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 5.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.6 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.8 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 1.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.2 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 3.4 | GO:0005741 | mitochondrial outer membrane(GO:0005741) organelle outer membrane(GO:0031968) |
| 0.0 | 4.7 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 3.2 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 1.4 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.4 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 12.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.6 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.9 | GO:0008021 | synaptic vesicle(GO:0008021) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 24.8 | 74.5 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 10.3 | 30.9 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 7.2 | 21.7 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 6.0 | 29.8 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 4.5 | 13.6 | GO:0000035 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 4.4 | 17.6 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 3.7 | 14.9 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 3.5 | 28.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 3.5 | 13.9 | GO:0031208 | POZ domain binding(GO:0031208) |
| 3.4 | 6.8 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 3.4 | 17.0 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 3.3 | 19.9 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 3.3 | 13.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 3.3 | 36.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 3.3 | 9.8 | GO:0001884 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 3.1 | 18.5 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 2.8 | 8.4 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 2.7 | 10.7 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 2.6 | 13.2 | GO:0030613 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 2.5 | 7.5 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 2.4 | 23.5 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 2.3 | 9.2 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) |
| 2.2 | 21.8 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 2.1 | 8.6 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 2.0 | 12.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 2.0 | 9.9 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 1.9 | 11.6 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 1.9 | 89.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 1.8 | 12.5 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 1.8 | 17.8 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 1.8 | 7.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 1.7 | 6.7 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 1.6 | 11.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 1.6 | 9.5 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 1.3 | 12.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 1.3 | 3.9 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 1.3 | 3.8 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 1.2 | 5.0 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 1.2 | 3.7 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 1.2 | 12.8 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 1.2 | 25.5 | GO:0000339 | RNA cap binding(GO:0000339) |
| 1.1 | 3.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 1.1 | 14.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 1.1 | 9.5 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 1.0 | 4.2 | GO:0030305 | beta-glucuronidase activity(GO:0004566) heparanase activity(GO:0030305) |
| 1.0 | 4.8 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.9 | 10.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.9 | 11.7 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.9 | 25.2 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.9 | 3.5 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.8 | 5.8 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.8 | 6.5 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.8 | 7.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.8 | 7.0 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.8 | 64.6 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.7 | 18.6 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.7 | 7.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.7 | 5.0 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.7 | 14.4 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.6 | 8.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.6 | 3.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.6 | 5.9 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.6 | 7.4 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.6 | 1.7 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.6 | 17.0 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.6 | 23.9 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.5 | 20.9 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.5 | 9.9 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.5 | 6.0 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.5 | 10.3 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.5 | 5.9 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.5 | 4.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.5 | 2.7 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.5 | 15.1 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.5 | 7.7 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.5 | 12.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.5 | 21.1 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.4 | 6.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.4 | 9.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.4 | 19.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.4 | 1.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.4 | 24.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.4 | 7.6 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.4 | 4.4 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.4 | 7.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.4 | 7.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.3 | 1.0 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.3 | 3.6 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.3 | 5.8 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.3 | 12.1 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.3 | 16.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.3 | 63.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.3 | 0.9 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.3 | 6.0 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.3 | 4.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.3 | 1.2 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.3 | 0.8 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.3 | 2.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.3 | 4.5 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.3 | 4.8 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.3 | 2.8 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.3 | 1.0 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.2 | 12.2 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.2 | 5.6 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 5.7 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 8.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.2 | 3.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 78.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.2 | 0.9 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.2 | 7.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.2 | 10.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.2 | 8.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.2 | 3.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 0.9 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.2 | 5.0 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.2 | 2.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 4.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 2.7 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 3.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 4.0 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 2.6 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.1 | 0.4 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 10.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 1.4 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 5.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 9.1 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 4.5 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.1 | 2.6 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 2.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 3.4 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.1 | 4.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 13.7 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 1.0 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 10.1 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 3.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 4.8 | GO:0008026 | ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.1 | 1.0 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 1.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 13.0 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 1.8 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.2 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 2.9 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 10.4 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 6.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 9.6 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.8 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 5.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 2.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 2.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 15.6 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 7.3 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.0 | 0.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.5 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 1.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 3.9 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.7 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 1.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.1 | GO:0019900 | kinase binding(GO:0019900) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.0 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.0 | GO:0004967 | glucagon receptor activity(GO:0004967) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 104.8 | PID BARD1 PATHWAY | BARD1 signaling events |
| 1.4 | 63.6 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.9 | 57.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.8 | 21.7 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.7 | 18.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.5 | 30.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.5 | 25.9 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.3 | 7.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.3 | 15.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.3 | 38.2 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.3 | 9.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.3 | 5.9 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.3 | 2.7 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.2 | 29.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 13.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 17.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.2 | 6.0 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.2 | 6.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.2 | 4.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.2 | 12.5 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.2 | 2.7 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 8.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 3.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 8.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 7.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 7.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 3.2 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 1.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 3.5 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 3.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 2.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 2.8 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.8 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 69.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 1.8 | 36.0 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 1.6 | 59.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 1.4 | 50.6 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 1.2 | 21.8 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 1.0 | 19.3 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 1.0 | 16.5 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.9 | 66.0 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.9 | 35.4 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.8 | 17.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.8 | 25.4 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.8 | 14.2 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.8 | 10.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.7 | 19.0 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.7 | 12.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.7 | 9.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.5 | 12.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.5 | 21.7 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.5 | 79.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.5 | 13.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.5 | 12.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.5 | 12.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.5 | 29.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.4 | 5.4 | REACTOME CDT1 ASSOCIATION WITH THE CDC6 ORC ORIGIN COMPLEX | Genes involved in CDT1 association with the CDC6:ORC:origin complex |
| 0.4 | 2.4 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.4 | 51.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.4 | 31.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.3 | 6.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.3 | 6.5 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.3 | 8.9 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.3 | 4.0 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.3 | 9.5 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.3 | 3.0 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.3 | 2.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.2 | 38.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.2 | 6.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 9.1 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.2 | 5.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 3.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 6.0 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.2 | 30.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 14.2 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.2 | 8.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 4.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.2 | 13.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.2 | 6.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 2.7 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 3.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 3.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 0.9 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.1 | 2.5 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 4.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 2.8 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 5.9 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 3.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 3.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 6.3 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 9.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 4.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 2.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.9 | REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | Genes involved in RNA Polymerase II Pre-transcription Events |
| 0.1 | 3.8 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 4.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 3.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.9 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 1.1 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 2.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 2.5 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |