Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
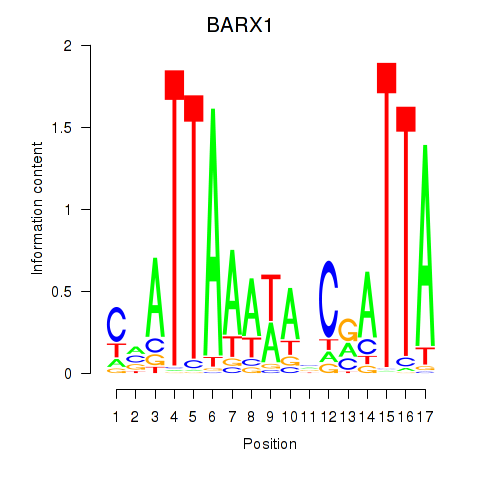
Results for BARX1
Z-value: 0.78
Transcription factors associated with BARX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BARX1
|
ENSG00000131668.9 | BARX homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BARX1 | hg19_v2_chr9_-_96717654_96717666 | -0.22 | 1.4e-03 | Click! |
Activity profile of BARX1 motif
Sorted Z-values of BARX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_159848807 | 21.91 |
ENST00000352433.5
|
PTTG1
|
pituitary tumor-transforming 1 |
| chr1_-_95391315 | 17.58 |
ENST00000545882.1
ENST00000415017.1 |
CNN3
|
calponin 3, acidic |
| chr12_-_10978957 | 16.94 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr7_+_107220660 | 16.84 |
ENST00000465919.1
ENST00000445771.2 ENST00000479917.1 ENST00000421217.1 ENST00000457837.1 |
BCAP29
|
B-cell receptor-associated protein 29 |
| chr1_-_197115818 | 16.64 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr2_-_106054952 | 16.64 |
ENST00000336660.5
ENST00000393352.3 ENST00000607522.1 |
FHL2
|
four and a half LIM domains 2 |
| chr16_-_66864806 | 15.97 |
ENST00000566336.1
ENST00000394074.2 ENST00000563185.2 ENST00000359087.4 ENST00000379463.2 ENST00000565535.1 ENST00000290810.3 |
NAE1
|
NEDD8 activating enzyme E1 subunit 1 |
| chr2_-_58468437 | 15.75 |
ENST00000403676.1
ENST00000427708.2 ENST00000403295.3 ENST00000446381.1 ENST00000417361.1 ENST00000233741.4 ENST00000402135.3 ENST00000540646.1 ENST00000449070.1 |
FANCL
|
Fanconi anemia, complementation group L |
| chr12_+_21207503 | 15.71 |
ENST00000545916.1
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr17_-_8113542 | 15.41 |
ENST00000578549.1
ENST00000535053.1 ENST00000582368.1 |
AURKB
|
aurora kinase B |
| chr3_-_149093499 | 15.02 |
ENST00000472441.1
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr12_-_52911718 | 14.56 |
ENST00000548409.1
|
KRT5
|
keratin 5 |
| chr10_+_62538089 | 14.56 |
ENST00000519078.2
ENST00000395284.3 ENST00000316629.4 |
CDK1
|
cyclin-dependent kinase 1 |
| chr12_+_104609550 | 14.09 |
ENST00000525566.1
ENST00000429002.2 |
TXNRD1
|
thioredoxin reductase 1 |
| chr5_+_140762268 | 13.84 |
ENST00000518325.1
|
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr3_-_64009658 | 13.60 |
ENST00000394431.2
|
PSMD6
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chr20_+_11898507 | 13.52 |
ENST00000378226.2
|
BTBD3
|
BTB (POZ) domain containing 3 |
| chr7_+_32996997 | 13.45 |
ENST00000242209.4
ENST00000538336.1 ENST00000538443.1 |
FKBP9
|
FK506 binding protein 9, 63 kDa |
| chrX_+_12809463 | 12.56 |
ENST00000380663.3
ENST00000380668.5 ENST00000398491.2 ENST00000489404.1 |
PRPS2
|
phosphoribosyl pyrophosphate synthetase 2 |
| chr8_-_141774467 | 12.52 |
ENST00000520151.1
ENST00000519024.1 ENST00000519465.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr4_+_88343952 | 12.41 |
ENST00000440591.2
|
NUDT9
|
nudix (nucleoside diphosphate linked moiety X)-type motif 9 |
| chrX_+_21958674 | 12.30 |
ENST00000404933.2
|
SMS
|
spermine synthase |
| chr15_-_72523924 | 12.15 |
ENST00000566809.1
ENST00000567087.1 ENST00000569050.1 ENST00000568883.1 |
PKM
|
pyruvate kinase, muscle |
| chr10_-_95242044 | 12.10 |
ENST00000371501.4
ENST00000371502.4 ENST00000371489.1 |
MYOF
|
myoferlin |
| chr2_+_130737223 | 11.90 |
ENST00000410061.2
|
RAB6C
|
RAB6C, member RAS oncogene family |
| chr3_+_160117087 | 11.87 |
ENST00000357388.3
|
SMC4
|
structural maintenance of chromosomes 4 |
| chr1_-_145826450 | 11.83 |
ENST00000462900.2
|
GPR89A
|
G protein-coupled receptor 89A |
| chr3_+_138340049 | 11.73 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr10_-_95241951 | 11.51 |
ENST00000358334.5
ENST00000359263.4 ENST00000371488.3 |
MYOF
|
myoferlin |
| chr14_+_55595762 | 11.17 |
ENST00000254301.9
|
LGALS3
|
lectin, galactoside-binding, soluble, 3 |
| chr1_-_21377447 | 10.99 |
ENST00000374937.3
ENST00000264211.8 |
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr2_+_231921574 | 10.83 |
ENST00000308696.6
ENST00000373635.4 ENST00000440838.1 ENST00000409643.1 |
PSMD1
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 |
| chr16_-_29934558 | 10.81 |
ENST00000568995.1
ENST00000566413.1 |
KCTD13
|
potassium channel tetramerization domain containing 13 |
| chr15_-_59949693 | 10.73 |
ENST00000396063.1
ENST00000396064.3 ENST00000484743.1 ENST00000559706.1 ENST00000396060.2 |
GTF2A2
|
general transcription factor IIA, 2, 12kDa |
| chrX_-_15511438 | 10.72 |
ENST00000380420.5
|
PIR
|
pirin (iron-binding nuclear protein) |
| chr4_+_169418195 | 10.61 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr1_-_150208363 | 10.54 |
ENST00000436748.2
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr3_-_185538849 | 10.47 |
ENST00000421047.2
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr3_-_186524234 | 10.45 |
ENST00000418288.1
ENST00000296273.2 |
RFC4
|
replication factor C (activator 1) 4, 37kDa |
| chr1_+_220267429 | 10.29 |
ENST00000366922.1
ENST00000302637.5 |
IARS2
|
isoleucyl-tRNA synthetase 2, mitochondrial |
| chr8_+_30244580 | 10.27 |
ENST00000523115.1
ENST00000519647.1 |
RBPMS
|
RNA binding protein with multiple splicing |
| chr6_+_63921399 | 10.05 |
ENST00000356170.3
|
FKBP1C
|
FK506 binding protein 1C |
| chr3_+_138340067 | 9.81 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr15_-_59949667 | 9.79 |
ENST00000396061.1
|
GTF2A2
|
general transcription factor IIA, 2, 12kDa |
| chr10_+_70748487 | 9.72 |
ENST00000361983.4
|
KIAA1279
|
KIAA1279 |
| chr22_-_19466732 | 9.71 |
ENST00000263202.10
ENST00000360834.4 |
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr19_-_55660561 | 9.51 |
ENST00000587758.1
ENST00000356783.5 ENST00000291901.8 ENST00000588426.1 ENST00000588147.1 ENST00000536926.1 ENST00000588981.1 |
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr3_-_123512688 | 9.39 |
ENST00000475616.1
|
MYLK
|
myosin light chain kinase |
| chr22_-_19466683 | 9.38 |
ENST00000399523.1
ENST00000421968.2 ENST00000447868.1 |
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr1_+_110546700 | 9.34 |
ENST00000359172.3
ENST00000393614.4 |
AHCYL1
|
adenosylhomocysteinase-like 1 |
| chr1_-_150208412 | 9.33 |
ENST00000532744.1
ENST00000369114.5 ENST00000369115.2 ENST00000369116.4 |
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr5_+_167913450 | 9.27 |
ENST00000231572.3
ENST00000538719.1 |
RARS
|
arginyl-tRNA synthetase |
| chr6_-_13814663 | 9.21 |
ENST00000359495.2
ENST00000379170.4 |
MCUR1
|
mitochondrial calcium uniporter regulator 1 |
| chr2_+_172544294 | 9.20 |
ENST00000358002.6
ENST00000435234.1 ENST00000443458.1 ENST00000412370.1 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr4_+_41540160 | 9.03 |
ENST00000503057.1
ENST00000511496.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr15_-_56757329 | 9.01 |
ENST00000260453.3
|
MNS1
|
meiosis-specific nuclear structural 1 |
| chr22_-_36220420 | 8.92 |
ENST00000473487.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr19_-_14945933 | 8.91 |
ENST00000322301.3
|
OR7A5
|
olfactory receptor, family 7, subfamily A, member 5 |
| chr6_-_131277510 | 8.85 |
ENST00000525193.1
ENST00000527659.1 |
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr13_+_48611665 | 8.84 |
ENST00000258662.2
|
NUDT15
|
nudix (nucleoside diphosphate linked moiety X)-type motif 15 |
| chr7_+_107220422 | 8.74 |
ENST00000005259.4
|
BCAP29
|
B-cell receptor-associated protein 29 |
| chr1_-_150208498 | 8.72 |
ENST00000314136.8
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr16_-_10652993 | 8.67 |
ENST00000536829.1
|
EMP2
|
epithelial membrane protein 2 |
| chr1_-_155904187 | 8.58 |
ENST00000368321.3
ENST00000368320.3 |
KIAA0907
|
KIAA0907 |
| chr1_-_150208291 | 8.54 |
ENST00000533654.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chrX_-_41449204 | 8.48 |
ENST00000378179.3
|
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr1_-_6420737 | 8.35 |
ENST00000541130.1
ENST00000377845.3 |
ACOT7
|
acyl-CoA thioesterase 7 |
| chr1_-_160492994 | 8.31 |
ENST00000368055.1
ENST00000368057.3 ENST00000368059.3 |
SLAMF6
|
SLAM family member 6 |
| chr9_+_108463234 | 8.23 |
ENST00000374688.1
|
TMEM38B
|
transmembrane protein 38B |
| chr17_+_18601299 | 8.20 |
ENST00000572555.1
ENST00000395902.3 ENST00000449552.2 |
TRIM16L
|
tripartite motif containing 16-like |
| chr16_-_21868978 | 8.17 |
ENST00000357370.5
ENST00000451409.1 ENST00000341400.7 ENST00000518761.4 |
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr6_-_114194483 | 8.14 |
ENST00000434296.2
|
RP1-249H1.4
|
RP1-249H1.4 |
| chr14_+_73563735 | 8.02 |
ENST00000532192.1
|
RBM25
|
RNA binding motif protein 25 |
| chr16_-_21436459 | 7.96 |
ENST00000448012.2
ENST00000504841.2 ENST00000419180.2 |
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr14_+_89060739 | 7.90 |
ENST00000318308.6
|
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr8_+_54764346 | 7.89 |
ENST00000297313.3
ENST00000344277.6 |
RGS20
|
regulator of G-protein signaling 20 |
| chrX_-_13835147 | 7.84 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chrX_-_134156502 | 7.84 |
ENST00000391440.1
|
FAM127C
|
family with sequence similarity 127, member C |
| chr12_-_71551652 | 7.79 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chr6_-_75960024 | 7.63 |
ENST00000370081.2
|
COX7A2
|
cytochrome c oxidase subunit VIIa polypeptide 2 (liver) |
| chr2_+_169312350 | 7.57 |
ENST00000305747.6
|
CERS6
|
ceramide synthase 6 |
| chr16_+_14802801 | 7.56 |
ENST00000526520.1
ENST00000531598.2 |
NPIPA3
|
nuclear pore complex interacting protein family, member A3 |
| chr22_+_19467261 | 7.37 |
ENST00000455750.1
ENST00000437685.2 ENST00000263201.1 ENST00000404724.3 |
CDC45
|
cell division cycle 45 |
| chr12_+_19282643 | 7.31 |
ENST00000317589.4
ENST00000355397.3 ENST00000359180.3 ENST00000309364.4 ENST00000540972.1 ENST00000429027.2 |
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr12_+_123237321 | 7.17 |
ENST00000280557.6
ENST00000455982.2 |
DENR
|
density-regulated protein |
| chr11_-_10828892 | 7.16 |
ENST00000525681.1
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr19_-_47349395 | 7.15 |
ENST00000597020.1
|
AP2S1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr13_+_98612446 | 7.14 |
ENST00000496368.1
ENST00000421861.2 ENST00000357602.3 |
IPO5
|
importin 5 |
| chr10_-_72648541 | 7.08 |
ENST00000299299.3
|
PCBD1
|
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha |
| chr7_-_105752971 | 7.08 |
ENST00000011473.2
|
SYPL1
|
synaptophysin-like 1 |
| chr20_+_34129770 | 7.02 |
ENST00000348547.2
ENST00000357394.4 ENST00000447986.1 ENST00000279052.6 ENST00000416206.1 ENST00000411577.1 ENST00000413587.1 |
ERGIC3
|
ERGIC and golgi 3 |
| chr8_-_87526561 | 7.01 |
ENST00000523911.1
|
RMDN1
|
regulator of microtubule dynamics 1 |
| chr16_+_22524844 | 7.01 |
ENST00000538606.1
ENST00000424340.1 ENST00000517539.1 ENST00000528249.1 |
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr9_-_69229650 | 6.87 |
ENST00000416428.1
|
CBWD6
|
COBW domain containing 6 |
| chr1_-_246729544 | 6.74 |
ENST00000544618.1
ENST00000366514.4 |
TFB2M
|
transcription factor B2, mitochondrial |
| chr6_-_56819385 | 6.69 |
ENST00000370754.5
ENST00000449297.2 |
DST
|
dystonin |
| chr5_-_55412774 | 6.67 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr1_-_43855444 | 6.66 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chr9_-_70465758 | 6.64 |
ENST00000489273.1
|
CBWD5
|
COBW domain containing 5 |
| chr2_+_237994519 | 6.61 |
ENST00000392008.2
ENST00000409334.1 ENST00000409629.1 |
COPS8
|
COP9 signalosome subunit 8 |
| chr5_+_72143988 | 6.56 |
ENST00000506351.2
|
TNPO1
|
transportin 1 |
| chr5_+_65222384 | 6.52 |
ENST00000380943.2
ENST00000416865.2 ENST00000380939.2 ENST00000380936.1 ENST00000380935.1 |
ERBB2IP
|
erbb2 interacting protein |
| chr16_-_29415350 | 6.52 |
ENST00000524087.1
|
NPIPB11
|
nuclear pore complex interacting protein family, member B11 |
| chr4_+_113568207 | 6.48 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr10_+_92631709 | 6.48 |
ENST00000413330.1
ENST00000277882.3 |
RPP30
|
ribonuclease P/MRP 30kDa subunit |
| chr12_+_20968608 | 6.47 |
ENST00000381541.3
ENST00000540229.1 ENST00000553473.1 ENST00000554957.1 |
LST3
SLCO1B3
SLCO1B7
|
Putative solute carrier organic anion transporter family member 1B7; Uncharacterized protein solute carrier organic anion transporter family, member 1B3 solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr2_+_242289502 | 6.40 |
ENST00000451310.1
|
SEPT2
|
septin 2 |
| chr3_+_8543393 | 6.39 |
ENST00000157600.3
ENST00000415597.1 ENST00000535732.1 |
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr8_+_132952112 | 6.36 |
ENST00000520362.1
ENST00000519656.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chrX_-_114253536 | 6.34 |
ENST00000371936.1
|
IL13RA2
|
interleukin 13 receptor, alpha 2 |
| chr20_-_47804894 | 6.33 |
ENST00000371828.3
ENST00000371856.2 ENST00000360426.4 ENST00000347458.5 ENST00000340954.7 ENST00000371802.1 ENST00000371792.1 ENST00000437404.2 |
STAU1
|
staufen double-stranded RNA binding protein 1 |
| chr2_-_175711133 | 6.29 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr1_+_73771844 | 6.26 |
ENST00000440762.1
ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1
|
RP4-598G3.1 |
| chrX_+_133733457 | 6.23 |
ENST00000440614.1
|
RP11-308B5.2
|
RP11-308B5.2 |
| chr3_-_176914238 | 6.16 |
ENST00000430069.1
ENST00000428970.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr7_+_16793160 | 6.12 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr14_+_55595960 | 6.08 |
ENST00000554715.1
|
LGALS3
|
lectin, galactoside-binding, soluble, 3 |
| chr15_-_66816853 | 6.02 |
ENST00000568588.1
|
RPL4
|
ribosomal protein L4 |
| chr1_-_54411255 | 5.91 |
ENST00000371377.3
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr16_-_29517141 | 5.86 |
ENST00000550665.1
|
RP11-231C14.4
|
Uncharacterized protein |
| chr12_+_20963632 | 5.78 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr8_-_101719159 | 5.73 |
ENST00000520868.1
ENST00000522658.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr1_-_21377383 | 5.69 |
ENST00000374935.3
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr14_-_75643296 | 5.68 |
ENST00000303575.4
|
TMED10
|
transmembrane emp24-like trafficking protein 10 (yeast) |
| chr2_+_11696464 | 5.66 |
ENST00000234142.5
|
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr20_-_45984401 | 5.65 |
ENST00000311275.7
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr14_-_50319758 | 5.62 |
ENST00000298310.5
|
NEMF
|
nuclear export mediator factor |
| chr4_-_17513851 | 5.60 |
ENST00000281243.5
|
QDPR
|
quinoid dihydropteridine reductase |
| chr12_-_121019165 | 5.56 |
ENST00000341039.2
ENST00000357500.4 |
POP5
|
processing of precursor 5, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr3_+_25831567 | 5.55 |
ENST00000280701.3
ENST00000420173.2 |
OXSM
|
3-oxoacyl-ACP synthase, mitochondrial |
| chr7_+_93551011 | 5.55 |
ENST00000248564.5
|
GNG11
|
guanine nucleotide binding protein (G protein), gamma 11 |
| chr8_+_87526645 | 5.52 |
ENST00000521271.1
ENST00000523072.1 ENST00000523001.1 ENST00000520814.1 ENST00000517771.1 |
CPNE3
|
copine III |
| chr5_-_43557791 | 5.51 |
ENST00000338972.4
ENST00000511321.1 ENST00000515338.1 |
PAIP1
|
poly(A) binding protein interacting protein 1 |
| chr15_-_52587945 | 5.50 |
ENST00000443683.2
ENST00000558479.1 ENST00000261839.7 |
MYO5C
|
myosin VC |
| chr8_-_109260897 | 5.38 |
ENST00000521297.1
ENST00000519030.1 ENST00000521440.1 ENST00000518345.1 ENST00000519627.1 ENST00000220849.5 |
EIF3E
|
eukaryotic translation initiation factor 3, subunit E |
| chr2_+_109335929 | 5.37 |
ENST00000283195.6
|
RANBP2
|
RAN binding protein 2 |
| chr6_-_2971429 | 5.33 |
ENST00000380529.1
|
SERPINB6
|
serpin peptidase inhibitor, clade B (ovalbumin), member 6 |
| chr7_-_105752651 | 5.30 |
ENST00000470347.1
ENST00000455385.2 |
SYPL1
|
synaptophysin-like 1 |
| chr5_-_16742330 | 5.26 |
ENST00000505695.1
ENST00000427430.2 |
MYO10
|
myosin X |
| chrX_-_114252193 | 5.26 |
ENST00000243213.1
|
IL13RA2
|
interleukin 13 receptor, alpha 2 |
| chr17_-_41132410 | 5.26 |
ENST00000409446.3
ENST00000453594.1 ENST00000409399.1 ENST00000421990.2 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr3_+_179280668 | 5.23 |
ENST00000429709.2
ENST00000450518.2 ENST00000392662.1 ENST00000490364.1 |
ACTL6A
|
actin-like 6A |
| chr11_-_102401469 | 5.23 |
ENST00000260227.4
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr22_+_19466980 | 5.21 |
ENST00000407835.1
ENST00000438587.1 |
CDC45
|
cell division cycle 45 |
| chr6_-_52859046 | 5.10 |
ENST00000457564.1
ENST00000541324.1 ENST00000370960.1 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr5_+_65222299 | 5.10 |
ENST00000284037.5
|
ERBB2IP
|
erbb2 interacting protein |
| chr20_+_56964253 | 5.10 |
ENST00000395802.3
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr7_-_72993033 | 5.09 |
ENST00000305632.5
|
TBL2
|
transducin (beta)-like 2 |
| chr11_+_20409070 | 5.05 |
ENST00000331079.6
|
PRMT3
|
protein arginine methyltransferase 3 |
| chr4_-_69817481 | 5.03 |
ENST00000251566.4
|
UGT2A3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr1_-_165668100 | 5.00 |
ENST00000354775.4
|
ALDH9A1
|
aldehyde dehydrogenase 9 family, member A1 |
| chr18_+_21529811 | 4.94 |
ENST00000588004.1
|
LAMA3
|
laminin, alpha 3 |
| chr11_+_57480046 | 4.94 |
ENST00000378312.4
ENST00000278422.4 |
TMX2
|
thioredoxin-related transmembrane protein 2 |
| chr6_+_37321823 | 4.92 |
ENST00000487950.1
ENST00000469731.1 |
RNF8
|
ring finger protein 8, E3 ubiquitin protein ligase |
| chr17_+_7155819 | 4.92 |
ENST00000570322.1
ENST00000576496.1 ENST00000574841.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr19_+_41281416 | 4.89 |
ENST00000597140.1
|
MIA
|
melanoma inhibitory activity |
| chr5_+_140797296 | 4.78 |
ENST00000398594.2
|
PCDHGB7
|
protocadherin gamma subfamily B, 7 |
| chr14_+_102276209 | 4.76 |
ENST00000445439.3
ENST00000334743.5 ENST00000557095.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr1_+_16083154 | 4.76 |
ENST00000375771.1
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr12_-_120884175 | 4.76 |
ENST00000546954.1
|
TRIAP1
|
TP53 regulated inhibitor of apoptosis 1 |
| chr9_-_21974820 | 4.74 |
ENST00000579122.1
ENST00000498124.1 |
CDKN2A
|
cyclin-dependent kinase inhibitor 2A |
| chr14_+_89060749 | 4.73 |
ENST00000555900.1
ENST00000406216.3 ENST00000557737.1 |
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr1_-_115292591 | 4.72 |
ENST00000438362.2
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr12_+_20963647 | 4.71 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr8_-_101718991 | 4.68 |
ENST00000517990.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr3_-_130465604 | 4.66 |
ENST00000356763.3
|
PIK3R4
|
phosphoinositide-3-kinase, regulatory subunit 4 |
| chr16_+_22501658 | 4.65 |
ENST00000415833.2
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr11_+_93479588 | 4.61 |
ENST00000526335.1
|
C11orf54
|
chromosome 11 open reading frame 54 |
| chr2_-_8723918 | 4.59 |
ENST00000454224.1
|
AC011747.4
|
AC011747.4 |
| chr5_+_140743859 | 4.57 |
ENST00000518069.1
|
PCDHGA5
|
protocadherin gamma subfamily A, 5 |
| chr6_+_123100620 | 4.57 |
ENST00000368444.3
|
FABP7
|
fatty acid binding protein 7, brain |
| chr12_+_50794592 | 4.53 |
ENST00000293618.8
ENST00000429001.3 ENST00000548174.1 ENST00000548697.1 ENST00000548993.1 ENST00000398473.2 ENST00000522085.1 ENST00000518444.1 ENST00000551886.1 |
LARP4
|
La ribonucleoprotein domain family, member 4 |
| chr5_-_141392538 | 4.51 |
ENST00000503794.1
ENST00000510194.1 ENST00000504424.1 ENST00000513454.1 ENST00000458112.2 ENST00000542860.1 ENST00000503229.1 ENST00000500692.2 ENST00000311337.6 ENST00000504139.1 ENST00000505689.1 |
GNPDA1
|
glucosamine-6-phosphate deaminase 1 |
| chrX_+_54466829 | 4.51 |
ENST00000375151.4
|
TSR2
|
TSR2, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr9_+_35161998 | 4.51 |
ENST00000396787.1
ENST00000378495.3 ENST00000378496.4 |
UNC13B
|
unc-13 homolog B (C. elegans) |
| chr5_-_9630463 | 4.50 |
ENST00000382492.2
|
TAS2R1
|
taste receptor, type 2, member 1 |
| chr7_+_107531580 | 4.48 |
ENST00000537148.1
ENST00000440410.1 ENST00000437604.2 |
DLD
|
dihydrolipoamide dehydrogenase |
| chr2_+_109237717 | 4.36 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr7_+_134576151 | 4.35 |
ENST00000393118.2
|
CALD1
|
caldesmon 1 |
| chr9_-_21202204 | 4.35 |
ENST00000239347.3
|
IFNA7
|
interferon, alpha 7 |
| chr2_-_74618964 | 4.34 |
ENST00000417090.1
ENST00000409868.1 |
DCTN1
|
dynactin 1 |
| chr7_+_134576317 | 4.32 |
ENST00000424922.1
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chr16_+_16434185 | 4.31 |
ENST00000524823.2
|
AC138969.4
|
Protein PKD1P1 |
| chr7_-_64023441 | 4.29 |
ENST00000309683.6
|
ZNF680
|
zinc finger protein 680 |
| chr5_-_125930929 | 4.29 |
ENST00000553117.1
ENST00000447989.2 ENST00000409134.3 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr8_+_107670064 | 4.27 |
ENST00000312046.6
|
OXR1
|
oxidation resistance 1 |
| chr1_-_236767779 | 4.20 |
ENST00000366579.1
ENST00000366582.3 ENST00000366581.2 |
HEATR1
|
HEAT repeat containing 1 |
| chr12_-_50290839 | 4.19 |
ENST00000552863.1
|
FAIM2
|
Fas apoptotic inhibitory molecule 2 |
| chrX_-_109590174 | 4.16 |
ENST00000372054.1
|
GNG5P2
|
guanine nucleotide binding protein (G protein), gamma 5 pseudogene 2 |
| chr11_-_86383650 | 4.13 |
ENST00000526944.1
ENST00000530335.1 ENST00000543262.1 ENST00000524826.1 |
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr19_-_53662257 | 4.11 |
ENST00000599096.1
ENST00000334197.7 ENST00000597183.1 ENST00000601804.1 ENST00000601469.2 ENST00000452676.2 |
ZNF347
|
zinc finger protein 347 |
| chr2_+_234621551 | 4.08 |
ENST00000608381.1
ENST00000373414.3 |
UGT1A1
UGT1A5
|
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr7_+_77469439 | 4.08 |
ENST00000450574.1
ENST00000416283.2 ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr11_+_44117260 | 4.05 |
ENST00000358681.4
|
EXT2
|
exostosin glycosyltransferase 2 |
| chr11_+_12766583 | 4.05 |
ENST00000361985.2
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr16_+_103816 | 3.95 |
ENST00000383018.3
ENST00000417493.1 |
SNRNP25
|
small nuclear ribonucleoprotein 25kDa (U11/U12) |
| chr4_+_37962018 | 3.95 |
ENST00000504686.1
|
PTTG2
|
pituitary tumor-transforming 2 |
| chr2_+_44001172 | 3.93 |
ENST00000260605.8
ENST00000406852.3 ENST00000443170.3 ENST00000398823.2 ENST00000605786.1 |
DYNC2LI1
|
dynein, cytoplasmic 2, light intermediate chain 1 |
| chr10_-_51130715 | 3.90 |
ENST00000402038.3
|
PARG
|
poly (ADP-ribose) glycohydrolase |
| chr15_+_79166065 | 3.88 |
ENST00000559690.1
ENST00000559158.1 |
MORF4L1
|
mortality factor 4 like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of BARX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 17.3 | GO:2000521 | negative regulation of immunological synapse formation(GO:2000521) |
| 5.1 | 15.4 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 5.1 | 5.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 4.2 | 12.6 | GO:0031938 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) regulation of chromatin silencing at telomere(GO:0031938) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 4.1 | 12.3 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 3.4 | 10.3 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 3.3 | 16.6 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 3.2 | 16.0 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 3.1 | 9.3 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 2.9 | 17.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 2.8 | 13.9 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 2.7 | 2.7 | GO:0060295 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 2.6 | 25.6 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 2.5 | 12.6 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 2.5 | 12.4 | GO:0009137 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 2.5 | 4.9 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 2.4 | 14.6 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 2.4 | 7.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 2.2 | 8.8 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 2.1 | 6.3 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 2.1 | 10.5 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 1.9 | 5.7 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 1.9 | 5.6 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 1.8 | 22.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 1.8 | 5.4 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 1.8 | 7.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 1.7 | 10.5 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 1.7 | 6.7 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 1.6 | 14.1 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 1.5 | 9.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 1.5 | 12.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 1.5 | 8.9 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 1.4 | 4.2 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 1.4 | 12.5 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 1.3 | 2.7 | GO:0006788 | heme oxidation(GO:0006788) |
| 1.3 | 9.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 1.3 | 11.6 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 1.2 | 7.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 1.2 | 9.4 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 1.2 | 3.5 | GO:1904395 | retinal rod cell differentiation(GO:0060221) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 1.2 | 4.7 | GO:0030242 | pexophagy(GO:0030242) |
| 1.2 | 10.4 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 1.1 | 14.9 | GO:0070986 | left/right axis specification(GO:0070986) |
| 1.1 | 4.5 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 1.1 | 4.5 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 1.1 | 3.4 | GO:0051257 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 1.1 | 9.5 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 1.0 | 8.4 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 1.0 | 2.9 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 1.0 | 8.7 | GO:0070836 | caveola assembly(GO:0070836) |
| 1.0 | 25.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.9 | 7.6 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.9 | 5.6 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.9 | 13.7 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.9 | 4.5 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.9 | 2.7 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.9 | 19.6 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.8 | 12.6 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.8 | 9.2 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.8 | 6.7 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.8 | 42.2 | GO:0043486 | histone exchange(GO:0043486) |
| 0.8 | 4.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.8 | 5.6 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.8 | 4.7 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.8 | 2.3 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.8 | 9.3 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.8 | 5.4 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.8 | 2.3 | GO:1901253 | regulation of intracellular transport of viral material(GO:1901252) negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.7 | 2.9 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.7 | 3.6 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.7 | 31.0 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.7 | 6.3 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.7 | 2.8 | GO:0035616 | maintenance of DNA methylation(GO:0010216) histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.7 | 7.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.7 | 2.1 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.7 | 4.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.7 | 8.9 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.7 | 19.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.7 | 2.0 | GO:0060613 | fat pad development(GO:0060613) |
| 0.6 | 30.2 | GO:2000816 | negative regulation of mitotic sister chromatid separation(GO:2000816) |
| 0.6 | 1.8 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.6 | 4.8 | GO:2001140 | regulation of phospholipid transport(GO:2001138) positive regulation of phospholipid transport(GO:2001140) |
| 0.6 | 1.1 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.6 | 8.5 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.6 | 5.6 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.6 | 8.3 | GO:0072540 | natural killer cell proliferation(GO:0001787) T-helper 17 cell lineage commitment(GO:0072540) |
| 0.5 | 10.3 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.5 | 11.9 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.5 | 4.8 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.5 | 2.5 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.5 | 5.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.5 | 2.5 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.5 | 2.5 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.5 | 2.5 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.5 | 3.9 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.5 | 1.4 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.5 | 2.4 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.5 | 1.4 | GO:1904154 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.5 | 8.5 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.5 | 2.3 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.5 | 2.3 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.4 | 23.7 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.4 | 4.7 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.4 | 1.7 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.4 | 5.4 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.4 | 6.6 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.4 | 3.7 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.4 | 18.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.4 | 10.8 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.4 | 7.2 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.4 | 2.4 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.4 | 1.5 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.4 | 6.5 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.4 | 3.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.4 | 1.4 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.3 | 2.4 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.3 | 9.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.3 | 25.4 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.3 | 2.3 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.3 | 16.4 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.3 | 2.6 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.3 | 4.8 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.3 | 3.7 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.3 | 2.2 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.3 | 1.5 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.3 | 0.9 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.3 | 4.2 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.3 | 3.3 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.3 | 2.3 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.3 | 26.1 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.3 | 16.3 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.3 | 7.0 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.3 | 1.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.3 | 4.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.2 | 0.7 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.2 | 3.2 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.2 | 1.2 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.2 | 11.8 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.2 | 5.7 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.2 | 1.9 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.2 | 0.7 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.2 | 4.7 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.2 | 3.1 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.2 | 0.7 | GO:1900158 | positive regulation of osteoclast proliferation(GO:0090290) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.2 | 4.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 2.0 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.2 | 4.0 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.2 | 6.3 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.2 | 9.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.2 | 0.9 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 0.9 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.2 | 1.4 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.2 | 0.5 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.2 | 1.0 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.2 | 5.1 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.2 | 1.1 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.2 | 1.9 | GO:1900016 | negative regulation of fibroblast migration(GO:0010764) negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.2 | 0.6 | GO:0051135 | NK T cell activation(GO:0051132) regulation of NK T cell activation(GO:0051133) positive regulation of NK T cell activation(GO:0051135) |
| 0.1 | 8.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 5.5 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 5.3 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 4.4 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.1 | 0.7 | GO:0021936 | regulation of cerebellar granule cell precursor proliferation(GO:0021936) positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.1 | 1.6 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.1 | 1.8 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 1.0 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.1 | 2.4 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 5.3 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.1 | 4.0 | GO:1902895 | positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.1 | 2.0 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.1 | 0.6 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 3.0 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.1 | 5.3 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 6.6 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 0.2 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.1 | 2.5 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 1.8 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 14.5 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 2.0 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 5.5 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 1.5 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 6.2 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 1.6 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 5.3 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.1 | 10.5 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.1 | 1.2 | GO:0019321 | pentose metabolic process(GO:0019321) neuron cellular homeostasis(GO:0070050) |
| 0.1 | 1.7 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.1 | 0.7 | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling(GO:0014066) |
| 0.1 | 1.1 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 11.3 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.1 | 10.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.6 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 6.7 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 1.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.5 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 2.8 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 0.3 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) |
| 0.1 | 2.4 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 0.9 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.1 | 1.3 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 1.0 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.1 | 4.5 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 4.5 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.1 | 2.2 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 4.7 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 6.4 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 3.8 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 2.9 | GO:0070125 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.0 | 3.8 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 4.4 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.3 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.6 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.6 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.2 | GO:0071428 | ribosomal subunit export from nucleus(GO:0000054) ribosomal large subunit export from nucleus(GO:0000055) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.0 | 1.4 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 1.6 | GO:0000165 | MAPK cascade(GO:0000165) |
| 0.0 | 1.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 1.3 | GO:0043067 | regulation of apoptotic process(GO:0042981) regulation of programmed cell death(GO:0043067) |
| 0.0 | 1.4 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.1 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.3 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 0.4 | GO:0006349 | regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.0 | 0.8 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 1.9 | GO:1903169 | regulation of calcium ion transmembrane transport(GO:1903169) |
| 0.0 | 2.6 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.1 | GO:1902172 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.0 | 0.3 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.5 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.7 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 2.5 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.5 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 1.5 | GO:0007162 | negative regulation of cell adhesion(GO:0007162) |
| 0.0 | 1.5 | GO:0007586 | digestion(GO:0007586) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.6 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 3.1 | 12.6 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 2.9 | 14.6 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 2.6 | 20.5 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 2.5 | 17.5 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 2.2 | 15.5 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 1.9 | 15.4 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 1.8 | 37.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 1.7 | 7.0 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 1.7 | 10.5 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 1.7 | 12.0 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 1.7 | 6.7 | GO:0031673 | H zone(GO:0031673) |
| 1.6 | 4.7 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 1.5 | 4.5 | GO:0044305 | calyx of Held(GO:0044305) |
| 1.5 | 4.4 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 1.4 | 4.2 | GO:0034455 | t-UTP complex(GO:0034455) |
| 1.2 | 12.2 | GO:0000796 | condensin complex(GO:0000796) |
| 1.1 | 3.4 | GO:0005715 | late recombination nodule(GO:0005715) |
| 1.1 | 23.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 1.0 | 7.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.9 | 2.7 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.9 | 5.4 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.8 | 4.0 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.8 | 25.1 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.7 | 3.7 | GO:0035061 | perichromatin fibrils(GO:0005726) interchromatin granule(GO:0035061) |
| 0.7 | 11.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.7 | 12.4 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.7 | 2.9 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.7 | 2.7 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.6 | 5.7 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.6 | 8.9 | GO:0008091 | spectrin(GO:0008091) |
| 0.6 | 4.9 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.6 | 2.4 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.6 | 2.3 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.5 | 10.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.5 | 2.7 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.5 | 8.5 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.5 | 7.9 | GO:0005802 | trans-Golgi network(GO:0005802) Golgi subcompartment(GO:0098791) |
| 0.5 | 4.7 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.5 | 16.7 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.5 | 5.7 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.5 | 7.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.5 | 2.0 | GO:0031417 | NatC complex(GO:0031417) |
| 0.5 | 9.1 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.5 | 2.4 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.5 | 2.8 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.5 | 6.4 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.4 | 9.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.4 | 4.7 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.4 | 12.0 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.4 | 9.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.4 | 1.6 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.4 | 2.3 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.4 | 7.2 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.4 | 1.9 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.4 | 2.9 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.4 | 2.2 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.3 | 15.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.3 | 8.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.3 | 9.2 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.3 | 8.0 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.3 | 2.5 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.3 | 2.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.3 | 6.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.3 | 17.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.3 | 5.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.3 | 1.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.3 | 14.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.3 | 0.8 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.3 | 13.0 | GO:0031430 | M band(GO:0031430) |
| 0.3 | 3.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.3 | 28.8 | GO:0005901 | caveola(GO:0005901) |
| 0.2 | 11.9 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 11.8 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.2 | 30.5 | GO:0042641 | actomyosin(GO:0042641) |
| 0.2 | 13.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.2 | 3.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 4.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 2.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.2 | 6.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.2 | 10.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 4.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.2 | 2.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 0.6 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.2 | 11.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.2 | 18.9 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.2 | 6.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 10.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 4.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.2 | 1.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.2 | 2.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.2 | 1.9 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 2.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 1.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 6.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 8.2 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.1 | 2.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 1.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 2.9 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 6.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 1.5 | GO:0038201 | TORC2 complex(GO:0031932) TOR complex(GO:0038201) |
| 0.1 | 10.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 4.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 31.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 4.8 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 1.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 7.0 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 13.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 15.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 12.0 | GO:0005840 | ribosome(GO:0005840) |
| 0.1 | 6.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 3.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 4.6 | GO:0031461 | cullin-RING ubiquitin ligase complex(GO:0031461) |
| 0.1 | 1.4 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 3.6 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 0.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 1.0 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.1 | 3.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 5.7 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.6 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.8 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 3.1 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 2.3 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 11.9 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 7.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.1 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 2.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 8.9 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 4.3 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.4 | GO:0012505 | endomembrane system(GO:0012505) |
| 0.0 | 0.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.8 | GO:0015629 | actin cytoskeleton(GO:0015629) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 14.1 | GO:0098626 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 4.1 | 12.4 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 3.4 | 10.3 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 3.1 | 9.3 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 3.0 | 12.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 2.9 | 8.8 | GO:0008413 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) NADH pyrophosphatase activity(GO:0035529) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 2.3 | 25.7 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 2.2 | 17.3 | GO:0019863 | IgE binding(GO:0019863) |
| 2.1 | 12.6 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 1.9 | 9.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 1.8 | 7.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 1.7 | 7.0 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 1.7 | 15.4 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 1.6 | 4.8 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 1.6 | 9.3 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 1.5 | 10.5 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 1.5 | 32.7 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 1.5 | 10.3 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 1.3 | 6.7 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 1.3 | 7.6 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 1.2 | 16.0 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 1.1 | 3.4 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 1.1 | 5.6 | GO:0070404 | NADH binding(GO:0070404) |
| 1.1 | 27.6 | GO:0008143 | poly(A) binding(GO:0008143) |
| 1.1 | 6.3 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) oxaloacetate decarboxylase activity(GO:0008948) |
| 1.0 | 2.9 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.9 | 2.7 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.9 | 8.9 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.9 | 9.7 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.9 | 3.5 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.8 | 8.4 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.8 | 8.4 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.8 | 3.3 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.8 | 2.4 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.8 | 19.7 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.8 | 12.6 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.8 | 10.8 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.8 | 2.3 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.8 | 14.6 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.8 | 16.7 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.8 | 12.0 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.7 | 12.5 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.7 | 20.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.7 | 7.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.7 | 2.9 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.7 | 36.1 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.7 | 4.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.6 | 18.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.6 | 8.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.6 | 12.0 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.6 | 2.9 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.6 | 5.3 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.6 | 5.3 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.6 | 23.3 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.6 | 4.5 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.6 | 1.7 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.5 | 5.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.5 | 5.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.5 | 2.3 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.4 | 17.1 | GO:0005215 | transporter activity(GO:0005215) |
| 0.4 | 1.3 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.4 | 3.4 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.4 | 4.5 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.4 | 19.1 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.4 | 11.2 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.4 | 9.3 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.4 | 11.8 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.4 | 10.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.4 | 8.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.4 | 2.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.3 | 22.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.3 | 1.7 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.3 | 7.2 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.3 | 8.4 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.3 | 6.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.3 | 7.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.3 | 1.8 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.3 | 2.3 | GO:0043559 | insulin binding(GO:0043559) |
| 0.3 | 4.0 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.3 | 8.9 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.3 | 1.4 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.3 | 4.7 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.3 | 1.0 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.3 | 4.8 | GO:0031005 | filamin binding(GO:0031005) |
| 0.3 | 4.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.2 | 6.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 0.7 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.2 | 0.9 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 16.6 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.2 | 0.7 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.2 | 2.8 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.2 | 4.0 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.2 | 1.6 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.2 | 3.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 3.2 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.2 | 4.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.2 | 5.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.2 | 3.6 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.2 | 2.2 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.2 | 17.2 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.2 | 14.5 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.2 | 0.6 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 9.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.2 | 1.8 | GO:0015386 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 13.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.2 | 1.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.2 | 5.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 7.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 0.8 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.2 | 4.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 2.0 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.2 | 0.5 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.2 | 2.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 2.4 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 5.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 1.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 1.5 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 0.9 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 20.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 2.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 2.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 1.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.9 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 21.0 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 4.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 4.7 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.1 | 1.0 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 2.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.7 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 19.8 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 9.8 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.1 | 1.5 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 31.6 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.1 | 15.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 6.7 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.1 | 2.4 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 3.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.6 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 3.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 9.8 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.8 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 1.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.3 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 3.7 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 10.8 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 10.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 1.1 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 3.2 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 1.0 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.9 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 1.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 2.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 5.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.7 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 1.2 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.0 | 3.8 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.6 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 1.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 5.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.5 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 14.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.6 | 33.0 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.5 | 34.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.4 | 5.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.4 | 12.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 12.5 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.3 | 12.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 22.0 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.2 | 5.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.2 | 23.6 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.2 | 6.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 4.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 4.7 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 2.9 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.2 | 7.9 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.2 | 7.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 4.9 | PID ATM PATHWAY | ATM pathway |
| 0.2 | 11.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 5.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 7.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 2.7 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 7.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 5.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 5.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 5.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 12.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 4.2 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.1 | 2.5 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 4.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 3.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 5.5 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 1.5 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 1.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 2.8 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 0.9 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 6.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 1.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 0.7 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 7.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 2.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 0.8 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 2.8 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 2.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 2.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.9 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.8 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 17.0 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 1.0 | 14.6 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 1.0 | 15.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.7 | 10.5 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.7 | 15.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.6 | 17.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.6 | 62.0 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.6 | 12.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.5 | 8.4 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.5 | 10.4 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.5 | 4.7 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.5 | 15.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.4 | 13.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.4 | 14.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.4 | 12.5 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.4 | 7.1 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.3 | 8.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.3 | 20.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.3 | 29.2 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.3 | 9.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.3 | 7.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.3 | 20.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.3 | 5.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.3 | 9.3 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.3 | 10.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.3 | 4.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.3 | 8.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.3 | 6.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 18.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.2 | 2.7 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.2 | 11.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 22.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.2 | 7.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 3.0 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 15.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.2 | 3.9 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.2 | 5.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 15.0 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.1 | 4.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 5.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 2.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 2.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.6 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.1 | 1.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 5.5 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 1.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 2.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 4.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 2.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.0 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 6.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 2.9 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 1.0 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 1.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 0.8 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.1 | 1.5 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 8.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 0.7 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 1.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.5 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 6.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.9 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.0 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 1.8 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.7 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 3.8 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |