Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
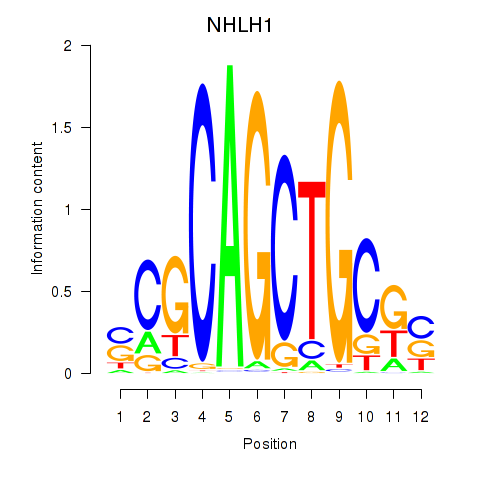
Results for NHLH1
Z-value: 0.71
Transcription factors associated with NHLH1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NHLH1
|
ENSG00000171786.5 | nescient helix-loop-helix 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NHLH1 | hg19_v2_chr1_+_160336851_160336868 | 0.24 | 3.6e-04 | Click! |
Activity profile of NHLH1 motif
Sorted Z-values of NHLH1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_23847267 | 64.41 |
ENST00000321728.7
|
PRKCB
|
protein kinase C, beta |
| chr16_+_23847339 | 47.08 |
ENST00000303531.7
|
PRKCB
|
protein kinase C, beta |
| chr14_+_90863327 | 29.62 |
ENST00000356978.4
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta) |
| chr10_+_105036909 | 27.30 |
ENST00000369849.4
|
INA
|
internexin neuronal intermediate filament protein, alpha |
| chr8_+_24771265 | 24.93 |
ENST00000518131.1
ENST00000437366.2 |
NEFM
|
neurofilament, medium polypeptide |
| chr8_+_24772455 | 22.87 |
ENST00000433454.2
|
NEFM
|
neurofilament, medium polypeptide |
| chr14_+_100150622 | 22.61 |
ENST00000261835.3
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1 |
| chr19_+_35521572 | 19.10 |
ENST00000262631.5
|
SCN1B
|
sodium channel, voltage-gated, type I, beta subunit |
| chr20_+_44036900 | 18.76 |
ENST00000443296.1
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr20_+_44036620 | 17.98 |
ENST00000372710.3
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr1_-_21948906 | 17.56 |
ENST00000374761.2
ENST00000599760.1 |
RAP1GAP
|
RAP1 GTPase activating protein |
| chr14_+_90863364 | 17.48 |
ENST00000447653.3
ENST00000553542.1 |
CALM1
|
calmodulin 1 (phosphorylase kinase, delta) |
| chr9_-_111929560 | 17.28 |
ENST00000561981.2
|
FRRS1L
|
ferric-chelate reductase 1-like |
| chr13_-_36705425 | 17.02 |
ENST00000255448.4
ENST00000360631.3 ENST00000379892.4 |
DCLK1
|
doublecortin-like kinase 1 |
| chr1_-_182360918 | 16.63 |
ENST00000339526.4
|
GLUL
|
glutamate-ammonia ligase |
| chr6_-_84418841 | 16.57 |
ENST00000369694.2
ENST00000195649.6 |
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr6_-_84418860 | 16.54 |
ENST00000521743.1
|
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr1_+_51701924 | 16.50 |
ENST00000242719.3
|
RNF11
|
ring finger protein 11 |
| chr6_-_84419101 | 16.23 |
ENST00000520302.1
ENST00000520213.1 ENST00000439399.2 ENST00000428679.2 ENST00000437520.1 |
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr20_+_37434329 | 15.87 |
ENST00000299824.1
ENST00000373331.2 |
PPP1R16B
|
protein phosphatase 1, regulatory subunit 16B |
| chr3_-_133614421 | 15.59 |
ENST00000543906.1
|
RAB6B
|
RAB6B, member RAS oncogene family |
| chr22_+_32340481 | 15.53 |
ENST00000397492.1
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr22_+_32340447 | 15.50 |
ENST00000248975.5
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr7_-_766879 | 15.20 |
ENST00000537384.1
ENST00000417852.1 |
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr16_+_6533380 | 15.19 |
ENST00000552089.1
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr19_+_35521616 | 15.03 |
ENST00000595652.1
|
SCN1B
|
sodium channel, voltage-gated, type I, beta subunit |
| chr17_-_56606664 | 15.00 |
ENST00000580844.1
|
SEPT4
|
septin 4 |
| chr17_-_56606705 | 14.95 |
ENST00000317268.3
|
SEPT4
|
septin 4 |
| chr17_-_56606639 | 14.86 |
ENST00000579371.1
|
SEPT4
|
septin 4 |
| chr4_-_102268484 | 14.78 |
ENST00000394853.4
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr1_-_3713042 | 14.49 |
ENST00000378251.1
|
LRRC47
|
leucine rich repeat containing 47 |
| chr1_-_85930823 | 14.39 |
ENST00000284031.8
ENST00000539042.1 |
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr4_-_102268628 | 14.13 |
ENST00000323055.6
ENST00000512215.1 ENST00000394854.3 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr16_+_330581 | 13.95 |
ENST00000219409.3
|
ARHGDIG
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr1_-_182360498 | 13.56 |
ENST00000417584.2
|
GLUL
|
glutamate-ammonia ligase |
| chr8_-_98290087 | 13.41 |
ENST00000322128.3
|
TSPYL5
|
TSPY-like 5 |
| chr11_+_17756279 | 13.13 |
ENST00000265969.6
|
KCNC1
|
potassium voltage-gated channel, Shaw-related subfamily, member 1 |
| chr1_+_33352036 | 13.02 |
ENST00000373467.3
|
HPCA
|
hippocalcin |
| chr3_+_167453493 | 12.96 |
ENST00000295777.5
ENST00000472747.2 |
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr3_-_133614597 | 12.93 |
ENST00000285208.4
ENST00000460865.3 |
RAB6B
|
RAB6B, member RAS oncogene family |
| chr5_+_156693091 | 12.70 |
ENST00000318218.6
ENST00000442283.2 ENST00000522463.1 ENST00000521420.1 |
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr5_+_156693159 | 12.48 |
ENST00000347377.6
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr18_-_5544241 | 12.39 |
ENST00000341928.2
ENST00000540638.2 |
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr19_+_47104493 | 12.14 |
ENST00000291295.9
ENST00000597743.1 |
CALM3
|
calmodulin 3 (phosphorylase kinase, delta) |
| chr11_-_66336060 | 12.03 |
ENST00000310325.5
|
CTSF
|
cathepsin F |
| chr12_-_6580094 | 11.85 |
ENST00000361716.3
|
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chrX_+_110187513 | 11.72 |
ENST00000446737.1
ENST00000425146.1 |
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3 |
| chr15_+_25200074 | 11.71 |
ENST00000390687.4
ENST00000584968.1 ENST00000346403.6 ENST00000554227.2 |
SNRPN
|
small nuclear ribonucleoprotein polypeptide N |
| chr13_-_45150392 | 11.66 |
ENST00000501704.2
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr4_+_154074217 | 11.50 |
ENST00000437508.2
|
TRIM2
|
tripartite motif containing 2 |
| chr17_-_79139817 | 11.41 |
ENST00000326724.4
|
AATK
|
apoptosis-associated tyrosine kinase |
| chr1_+_50575292 | 11.37 |
ENST00000371821.1
ENST00000371819.1 |
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr19_-_49944806 | 11.12 |
ENST00000221485.3
|
SLC17A7
|
solute carrier family 17 (vesicular glutamate transporter), member 7 |
| chr1_-_182361327 | 11.11 |
ENST00000331872.6
ENST00000311223.5 |
GLUL
|
glutamate-ammonia ligase |
| chr1_-_177134024 | 11.02 |
ENST00000367654.3
|
ASTN1
|
astrotactin 1 |
| chr15_+_25200108 | 11.02 |
ENST00000577949.1
ENST00000338094.6 ENST00000338327.4 ENST00000579070.1 ENST00000577565.1 |
SNURF
SNRPN
|
SNRPN upstream reading frame protein small nuclear ribonucleoprotein polypeptide N |
| chr2_-_86564776 | 10.89 |
ENST00000165698.5
ENST00000541910.1 ENST00000535845.1 |
REEP1
|
receptor accessory protein 1 |
| chr3_-_33700933 | 10.61 |
ENST00000480013.1
|
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr17_-_27278304 | 10.58 |
ENST00000577226.1
|
PHF12
|
PHD finger protein 12 |
| chr16_+_6533729 | 10.35 |
ENST00000551752.1
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr16_+_1756162 | 10.27 |
ENST00000250894.4
ENST00000356010.5 |
MAPK8IP3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr11_-_66725837 | 10.11 |
ENST00000393958.2
ENST00000393960.1 ENST00000524491.1 ENST00000355677.3 |
PC
|
pyruvate carboxylase |
| chr12_-_90102594 | 10.08 |
ENST00000428670.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr13_+_35516390 | 10.05 |
ENST00000540320.1
ENST00000400445.3 ENST00000310336.4 |
NBEA
|
neurobeachin |
| chr6_+_30848557 | 9.90 |
ENST00000460944.2
ENST00000324771.8 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr5_+_71403061 | 9.81 |
ENST00000512974.1
ENST00000296755.7 |
MAP1B
|
microtubule-associated protein 1B |
| chr3_-_47619623 | 9.77 |
ENST00000456150.1
|
CSPG5
|
chondroitin sulfate proteoglycan 5 (neuroglycan C) |
| chr11_-_695162 | 9.64 |
ENST00000338675.6
|
DEAF1
|
DEAF1 transcription factor |
| chr6_-_167040731 | 9.42 |
ENST00000265678.4
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr15_+_43803143 | 9.34 |
ENST00000382031.1
|
MAP1A
|
microtubule-associated protein 1A |
| chr1_-_177133818 | 9.24 |
ENST00000424564.2
ENST00000361833.2 |
ASTN1
|
astrotactin 1 |
| chr4_-_186697044 | 9.22 |
ENST00000437304.2
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr16_-_29910365 | 9.20 |
ENST00000346932.5
ENST00000350527.3 ENST00000537485.1 ENST00000568380.1 |
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
| chr1_+_87797351 | 9.19 |
ENST00000370542.1
|
LMO4
|
LIM domain only 4 |
| chr14_-_27066960 | 9.16 |
ENST00000539517.2
|
NOVA1
|
neuro-oncological ventral antigen 1 |
| chr20_+_57430162 | 9.15 |
ENST00000450130.1
ENST00000349036.3 ENST00000423897.1 |
GNAS
|
GNAS complex locus |
| chr6_-_3157760 | 9.00 |
ENST00000333628.3
|
TUBB2A
|
tubulin, beta 2A class IIa |
| chr8_+_86376081 | 8.64 |
ENST00000285379.5
|
CA2
|
carbonic anhydrase II |
| chr12_-_6579808 | 8.63 |
ENST00000535180.1
ENST00000400911.3 |
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr22_+_19705928 | 8.55 |
ENST00000383045.3
ENST00000438754.2 |
SEPT5
|
septin 5 |
| chr6_+_29624862 | 8.46 |
ENST00000376894.4
|
MOG
|
myelin oligodendrocyte glycoprotein |
| chr2_-_11810284 | 8.44 |
ENST00000306928.5
|
NTSR2
|
neurotensin receptor 2 |
| chr19_+_47104553 | 8.20 |
ENST00000598871.1
ENST00000594523.1 |
CALM3
|
calmodulin 3 (phosphorylase kinase, delta) |
| chr6_+_29624758 | 8.14 |
ENST00000376917.3
ENST00000376902.3 ENST00000533330.2 ENST00000376888.2 |
MOG
|
myelin oligodendrocyte glycoprotein |
| chr3_+_35681081 | 8.00 |
ENST00000428373.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr6_+_29624898 | 7.99 |
ENST00000396704.3
ENST00000483013.1 ENST00000490427.1 ENST00000416766.2 ENST00000376891.4 ENST00000376898.3 ENST00000396701.2 ENST00000494692.1 ENST00000431798.2 |
MOG
|
myelin oligodendrocyte glycoprotein |
| chr8_+_41348173 | 7.69 |
ENST00000357743.4
|
GOLGA7
|
golgin A7 |
| chr20_+_20348740 | 7.50 |
ENST00000310227.1
|
INSM1
|
insulinoma-associated 1 |
| chr10_-_127511790 | 7.49 |
ENST00000368797.4
ENST00000420761.1 |
UROS
|
uroporphyrinogen III synthase |
| chr14_-_23772032 | 7.43 |
ENST00000452015.4
|
PPP1R3E
|
protein phosphatase 1, regulatory subunit 3E |
| chr3_+_50712672 | 7.37 |
ENST00000266037.9
|
DOCK3
|
dedicator of cytokinesis 3 |
| chr13_+_88324870 | 7.34 |
ENST00000325089.6
|
SLITRK5
|
SLIT and NTRK-like family, member 5 |
| chr7_-_767249 | 7.34 |
ENST00000403562.1
|
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr2_-_73511559 | 7.22 |
ENST00000521871.1
|
FBXO41
|
F-box protein 41 |
| chr14_-_65438865 | 7.14 |
ENST00000267512.5
|
RAB15
|
RAB15, member RAS oncogene family |
| chr20_+_34043085 | 7.07 |
ENST00000397527.1
ENST00000342580.4 |
CEP250
|
centrosomal protein 250kDa |
| chr8_+_41348072 | 7.05 |
ENST00000405786.2
|
GOLGA7
|
golgin A7 |
| chr12_+_121088291 | 7.05 |
ENST00000351200.2
|
CABP1
|
calcium binding protein 1 |
| chr1_+_174846570 | 6.97 |
ENST00000392064.2
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr11_+_71238313 | 6.92 |
ENST00000398536.4
|
KRTAP5-7
|
keratin associated protein 5-7 |
| chr14_+_102027688 | 6.85 |
ENST00000510508.4
ENST00000359323.3 |
DIO3
|
deiodinase, iodothyronine, type III |
| chr11_-_35441524 | 6.77 |
ENST00000395750.1
ENST00000449068.1 |
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr19_+_4304632 | 6.72 |
ENST00000597590.1
|
FSD1
|
fibronectin type III and SPRY domain containing 1 |
| chr19_+_4304585 | 6.67 |
ENST00000221856.6
|
FSD1
|
fibronectin type III and SPRY domain containing 1 |
| chr22_+_40390930 | 6.58 |
ENST00000333407.6
|
FAM83F
|
family with sequence similarity 83, member F |
| chr16_+_330448 | 6.51 |
ENST00000447871.1
|
ARHGDIG
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr9_-_79307096 | 6.50 |
ENST00000376717.2
ENST00000223609.6 ENST00000443509.2 |
PRUNE2
|
prune homolog 2 (Drosophila) |
| chr3_-_194991876 | 6.33 |
ENST00000310380.6
|
XXYLT1
|
xyloside xylosyltransferase 1 |
| chr7_+_45613958 | 6.19 |
ENST00000297323.7
|
ADCY1
|
adenylate cyclase 1 (brain) |
| chr7_+_110731062 | 6.15 |
ENST00000308478.5
ENST00000451085.1 ENST00000422987.3 ENST00000421101.1 |
LRRN3
|
leucine rich repeat neuronal 3 |
| chr17_-_56595196 | 6.14 |
ENST00000579921.1
ENST00000579925.1 ENST00000323456.5 |
MTMR4
|
myotubularin related protein 4 |
| chr16_-_74808710 | 6.14 |
ENST00000219368.3
ENST00000544337.1 |
FA2H
|
fatty acid 2-hydroxylase |
| chrX_-_68385354 | 6.07 |
ENST00000361478.1
|
PJA1
|
praja ring finger 1, E3 ubiquitin protein ligase |
| chr19_+_10217364 | 5.99 |
ENST00000430370.1
|
PPAN
|
peter pan homolog (Drosophila) |
| chr2_+_17935383 | 5.87 |
ENST00000524465.1
ENST00000381254.2 ENST00000532257.1 |
GEN1
|
GEN1 Holliday junction 5' flap endonuclease |
| chr10_-_81205373 | 5.86 |
ENST00000372336.3
|
ZCCHC24
|
zinc finger, CCHC domain containing 24 |
| chrX_-_68385274 | 5.85 |
ENST00000374584.3
ENST00000590146.1 |
PJA1
|
praja ring finger 1, E3 ubiquitin protein ligase |
| chr5_+_139493665 | 5.74 |
ENST00000331327.3
|
PURA
|
purine-rich element binding protein A |
| chr16_+_22825475 | 5.72 |
ENST00000261374.3
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2 |
| chr1_+_233749739 | 5.68 |
ENST00000366621.3
|
KCNK1
|
potassium channel, subfamily K, member 1 |
| chr6_-_146056341 | 5.63 |
ENST00000435470.1
|
EPM2A
|
epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) |
| chr10_-_103347883 | 5.56 |
ENST00000339310.3
ENST00000370158.3 ENST00000299206.4 ENST00000456836.2 ENST00000413344.1 ENST00000429502.1 ENST00000430045.1 ENST00000370172.1 ENST00000436284.2 ENST00000370162.3 |
POLL
|
polymerase (DNA directed), lambda |
| chr11_+_125774258 | 5.50 |
ENST00000263576.6
|
DDX25
|
DEAD (Asp-Glu-Ala-Asp) box helicase 25 |
| chr14_-_65439132 | 5.49 |
ENST00000533601.2
|
RAB15
|
RAB15, member RAS oncogene family |
| chr4_-_186696425 | 5.42 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr11_-_35441597 | 5.36 |
ENST00000395753.1
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr19_-_14316980 | 5.31 |
ENST00000361434.3
ENST00000340736.6 |
LPHN1
|
latrophilin 1 |
| chr1_+_226411319 | 5.30 |
ENST00000542034.1
ENST00000366810.5 |
MIXL1
|
Mix paired-like homeobox |
| chr1_+_89990431 | 5.11 |
ENST00000330947.2
ENST00000358200.4 |
LRRC8B
|
leucine rich repeat containing 8 family, member B |
| chr1_-_233431458 | 5.06 |
ENST00000258229.9
ENST00000430153.1 |
PCNXL2
|
pecanex-like 2 (Drosophila) |
| chr10_+_1102721 | 5.03 |
ENST00000263150.4
|
WDR37
|
WD repeat domain 37 |
| chr15_+_74833518 | 4.93 |
ENST00000346246.5
|
ARID3B
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr1_-_84464780 | 4.86 |
ENST00000260505.8
|
TTLL7
|
tubulin tyrosine ligase-like family, member 7 |
| chr10_+_18429671 | 4.84 |
ENST00000282343.8
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr6_+_126112001 | 4.81 |
ENST00000392477.2
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr21_+_34697258 | 4.77 |
ENST00000442071.1
ENST00000442357.2 |
IFNAR1
|
interferon (alpha, beta and omega) receptor 1 |
| chr2_+_17935119 | 4.76 |
ENST00000317402.7
|
GEN1
|
GEN1 Holliday junction 5' flap endonuclease |
| chr7_+_99613212 | 4.72 |
ENST00000426572.1
ENST00000535170.1 |
ZKSCAN1
|
zinc finger with KRAB and SCAN domains 1 |
| chr7_+_72848092 | 4.71 |
ENST00000344575.3
|
FZD9
|
frizzled family receptor 9 |
| chr19_-_47164386 | 4.65 |
ENST00000391916.2
ENST00000410105.2 |
DACT3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr20_+_49575342 | 4.58 |
ENST00000244051.1
|
MOCS3
|
molybdenum cofactor synthesis 3 |
| chr2_+_11052054 | 4.56 |
ENST00000295082.1
|
KCNF1
|
potassium voltage-gated channel, subfamily F, member 1 |
| chr6_+_30851840 | 4.56 |
ENST00000511510.1
ENST00000376569.3 ENST00000376575.3 ENST00000376570.4 ENST00000446312.1 ENST00000504927.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr17_-_73401728 | 4.43 |
ENST00000316804.5
ENST00000316615.5 |
GRB2
|
growth factor receptor-bound protein 2 |
| chr5_+_118788182 | 4.43 |
ENST00000515320.1
ENST00000510025.1 |
HSD17B4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr21_+_40752170 | 4.41 |
ENST00000333781.5
ENST00000541890.1 |
WRB
|
tryptophan rich basic protein |
| chr17_-_31620006 | 4.39 |
ENST00000225823.2
|
ASIC2
|
acid-sensing (proton-gated) ion channel 2 |
| chrX_-_107018969 | 4.35 |
ENST00000372383.4
|
TSC22D3
|
TSC22 domain family, member 3 |
| chr19_+_5904866 | 4.33 |
ENST00000339485.3
|
VMAC
|
vimentin-type intermediate filament associated coiled-coil protein |
| chr12_+_107168342 | 4.33 |
ENST00000392837.4
|
RIC8B
|
RIC8 guanine nucleotide exchange factor B |
| chr5_-_19988339 | 4.29 |
ENST00000382275.1
|
CDH18
|
cadherin 18, type 2 |
| chr19_+_12949251 | 4.28 |
ENST00000251472.4
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chr2_+_65283506 | 4.27 |
ENST00000377990.2
|
CEP68
|
centrosomal protein 68kDa |
| chr12_+_57984965 | 4.27 |
ENST00000540759.2
ENST00000551772.1 ENST00000550465.1 ENST00000354947.5 |
PIP4K2C
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma |
| chr17_+_47653178 | 4.26 |
ENST00000328741.5
|
NXPH3
|
neurexophilin 3 |
| chr1_+_44115814 | 4.23 |
ENST00000372396.3
|
KDM4A
|
lysine (K)-specific demethylase 4A |
| chr2_-_230135937 | 4.21 |
ENST00000392054.3
ENST00000409462.1 ENST00000392055.3 |
PID1
|
phosphotyrosine interaction domain containing 1 |
| chr22_-_20850070 | 4.18 |
ENST00000440659.2
ENST00000458248.1 ENST00000443285.1 ENST00000444967.1 ENST00000451553.1 ENST00000431430.1 |
KLHL22
|
kelch-like family member 22 |
| chr6_+_37012607 | 4.11 |
ENST00000423336.1
|
COX6A1P2
|
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
| chr5_+_118788261 | 4.05 |
ENST00000504811.1
|
HSD17B4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr14_-_45603657 | 4.05 |
ENST00000396062.3
|
FKBP3
|
FK506 binding protein 3, 25kDa |
| chr1_+_24969755 | 4.05 |
ENST00000447431.2
ENST00000374389.4 |
SRRM1
|
serine/arginine repetitive matrix 1 |
| chr2_+_231902193 | 4.03 |
ENST00000373640.4
|
C2orf72
|
chromosome 2 open reading frame 72 |
| chr4_+_62066941 | 4.00 |
ENST00000512091.2
|
LPHN3
|
latrophilin 3 |
| chr1_+_156124162 | 3.99 |
ENST00000368282.1
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr16_+_19222479 | 3.97 |
ENST00000568433.1
|
SYT17
|
synaptotagmin XVII |
| chr7_+_99613195 | 3.95 |
ENST00000324306.6
|
ZKSCAN1
|
zinc finger with KRAB and SCAN domains 1 |
| chr6_-_154831779 | 3.92 |
ENST00000607772.1
|
CNKSR3
|
CNKSR family member 3 |
| chr4_+_176986978 | 3.92 |
ENST00000508596.1
ENST00000393643.2 |
WDR17
|
WD repeat domain 17 |
| chr5_-_133747589 | 3.88 |
ENST00000458198.2
|
CDKN2AIPNL
|
CDKN2A interacting protein N-terminal like |
| chr12_-_120554622 | 3.86 |
ENST00000229340.5
|
RAB35
|
RAB35, member RAS oncogene family |
| chr19_-_45996465 | 3.84 |
ENST00000430715.2
|
RTN2
|
reticulon 2 |
| chr2_+_8822113 | 3.84 |
ENST00000396290.1
ENST00000331129.3 |
ID2
|
inhibitor of DNA binding 2, dominant negative helix-loop-helix protein |
| chr1_-_153919128 | 3.82 |
ENST00000361217.4
|
DENND4B
|
DENN/MADD domain containing 4B |
| chr1_+_202317815 | 3.81 |
ENST00000608999.1
ENST00000336894.4 ENST00000480184.1 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr10_-_98945677 | 3.80 |
ENST00000266058.4
ENST00000371041.3 |
SLIT1
|
slit homolog 1 (Drosophila) |
| chr10_+_92980517 | 3.79 |
ENST00000336126.5
|
PCGF5
|
polycomb group ring finger 5 |
| chr8_-_125740514 | 3.78 |
ENST00000325064.5
ENST00000518547.1 |
MTSS1
|
metastasis suppressor 1 |
| chr1_-_237167718 | 3.78 |
ENST00000464121.2
|
MT1HL1
|
metallothionein 1H-like 1 |
| chr6_+_26156551 | 3.76 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr19_-_54327542 | 3.76 |
ENST00000391775.3
ENST00000324134.6 ENST00000535162.1 ENST00000351894.4 ENST00000354278.3 ENST00000391773.1 ENST00000345770.5 ENST00000391772.1 |
NLRP12
|
NLR family, pyrin domain containing 12 |
| chr5_-_16936340 | 3.76 |
ENST00000507288.1
ENST00000513610.1 |
MYO10
|
myosin X |
| chr2_-_202316260 | 3.74 |
ENST00000332624.3
|
TRAK2
|
trafficking protein, kinesin binding 2 |
| chr17_-_73401567 | 3.69 |
ENST00000392562.1
|
GRB2
|
growth factor receptor-bound protein 2 |
| chr2_-_9563319 | 3.68 |
ENST00000497105.1
ENST00000360635.3 ENST00000359712.3 |
ITGB1BP1
|
integrin beta 1 binding protein 1 |
| chr9_+_125703282 | 3.65 |
ENST00000373647.4
ENST00000402311.1 |
RABGAP1
|
RAB GTPase activating protein 1 |
| chr14_-_104313824 | 3.55 |
ENST00000553739.1
ENST00000202556.9 |
PPP1R13B
|
protein phosphatase 1, regulatory subunit 13B |
| chr10_+_18689637 | 3.54 |
ENST00000377315.4
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr2_+_236578248 | 3.51 |
ENST00000409538.1
|
AGAP1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr1_-_85156090 | 3.51 |
ENST00000605755.1
ENST00000437941.2 |
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr2_+_65283529 | 3.51 |
ENST00000546106.1
ENST00000537589.1 ENST00000260569.4 |
CEP68
|
centrosomal protein 68kDa |
| chr5_+_118788433 | 3.50 |
ENST00000414835.2
|
HSD17B4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr19_-_460996 | 3.47 |
ENST00000264554.6
|
SHC2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr5_+_118788138 | 3.47 |
ENST00000256216.6
|
HSD17B4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr12_+_122064398 | 3.45 |
ENST00000330079.7
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1 |
| chr10_+_12391481 | 3.43 |
ENST00000378847.3
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID |
| chr20_+_8112824 | 3.40 |
ENST00000378641.3
|
PLCB1
|
phospholipase C, beta 1 (phosphoinositide-specific) |
| chr19_-_3061397 | 3.39 |
ENST00000586839.1
|
AES
|
amino-terminal enhancer of split |
| chr19_+_7598890 | 3.39 |
ENST00000221249.6
ENST00000601668.1 ENST00000601001.1 |
PNPLA6
|
patatin-like phospholipase domain containing 6 |
| chr14_+_24583836 | 3.35 |
ENST00000559115.1
ENST00000558215.1 ENST00000557810.1 ENST00000561375.1 ENST00000446197.3 ENST00000559796.1 ENST00000560713.1 ENST00000560901.1 ENST00000559382.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NHLH1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 37.2 | 111.5 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 15.9 | 47.8 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 11.4 | 34.1 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 10.3 | 41.3 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 9.6 | 28.9 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 9.0 | 44.8 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 5.9 | 17.6 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 5.3 | 15.9 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 3.9 | 27.3 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 3.7 | 11.1 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 3.6 | 14.5 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 3.5 | 10.6 | GO:0071139 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 3.4 | 10.1 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 3.1 | 25.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 2.9 | 14.7 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 2.9 | 8.6 | GO:0042938 | dipeptide transport(GO:0042938) |
| 2.6 | 13.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 2.6 | 13.0 | GO:1902075 | cellular response to salt(GO:1902075) |
| 2.6 | 15.4 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 2.5 | 7.5 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 2.3 | 22.6 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 2.0 | 10.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 1.9 | 9.4 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 1.9 | 7.5 | GO:0046502 | uroporphyrinogen III biosynthetic process(GO:0006780) uroporphyrinogen III metabolic process(GO:0046502) |
| 1.8 | 12.4 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 1.7 | 49.2 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 1.6 | 14.5 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 1.6 | 4.7 | GO:1990523 | negative regulation of neuromuscular junction development(GO:1904397) bone regeneration(GO:1990523) |
| 1.6 | 31.0 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 1.5 | 4.6 | GO:0018307 | enzyme active site formation(GO:0018307) tRNA thio-modification(GO:0034227) |
| 1.5 | 12.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 1.4 | 22.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 1.4 | 4.2 | GO:0090298 | negative regulation of mitochondrial DNA replication(GO:0090298) negative regulation of mitochondrial DNA metabolic process(GO:1901859) |
| 1.4 | 8.4 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 1.3 | 10.6 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 1.3 | 9.1 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 1.2 | 3.7 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 1.2 | 5.0 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 1.2 | 3.7 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 1.2 | 4.7 | GO:0031179 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 1.1 | 3.4 | GO:0090427 | activation of meiosis(GO:0090427) |
| 1.1 | 4.5 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 1.1 | 14.4 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 1.1 | 21.3 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 1.0 | 6.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 1.0 | 5.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 1.0 | 2.9 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 1.0 | 6.9 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 1.0 | 2.9 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.9 | 3.8 | GO:0032661 | Toll signaling pathway(GO:0008063) regulation of interleukin-18 production(GO:0032661) |
| 0.9 | 2.8 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.9 | 5.6 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.9 | 8.0 | GO:0009408 | response to heat(GO:0009408) cellular response to heat(GO:0034605) |
| 0.8 | 3.3 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) negative regulation of protein kinase C signaling(GO:0090038) |
| 0.8 | 9.2 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.8 | 3.3 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.8 | 2.5 | GO:0051054 | positive regulation of DNA metabolic process(GO:0051054) |
| 0.8 | 2.3 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.8 | 3.8 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.8 | 9.8 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.8 | 20.3 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.7 | 4.4 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.7 | 5.1 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.7 | 2.2 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.7 | 11.4 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.7 | 5.7 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.7 | 6.1 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.7 | 17.0 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.7 | 5.3 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.6 | 1.9 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.6 | 3.8 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.6 | 3.8 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.6 | 4.4 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.6 | 2.5 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.6 | 8.1 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.6 | 6.1 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.6 | 4.2 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.6 | 7.8 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.6 | 1.7 | GO:0090611 | mitotic cytokinesis checkpoint(GO:0044878) ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.6 | 5.7 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.6 | 2.8 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.6 | 3.4 | GO:0071233 | cellular response to leucine(GO:0071233) cellular response to leucine starvation(GO:1990253) |
| 0.6 | 2.2 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.5 | 2.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.5 | 2.7 | GO:0036115 | fatty-acyl-CoA catabolic process(GO:0036115) malonyl-CoA metabolic process(GO:2001293) |
| 0.5 | 3.3 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.5 | 9.6 | GO:0033599 | regulation of mammary gland epithelial cell proliferation(GO:0033599) |
| 0.5 | 4.6 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.5 | 6.2 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.5 | 1.0 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.5 | 1.4 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.5 | 3.8 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.5 | 3.7 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.5 | 5.5 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.4 | 12.5 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.4 | 4.3 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.4 | 2.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.4 | 1.7 | GO:1902949 | positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.4 | 1.2 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.4 | 6.0 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.4 | 9.9 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.4 | 0.8 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.4 | 1.2 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.4 | 1.1 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.3 | 7.3 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.3 | 1.4 | GO:0060023 | soft palate development(GO:0060023) |
| 0.3 | 1.4 | GO:0032811 | negative regulation of epinephrine secretion(GO:0032811) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.3 | 24.3 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.3 | 11.1 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.3 | 3.3 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.3 | 4.6 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.3 | 2.0 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.3 | 1.0 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.3 | 1.3 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.3 | 3.4 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.3 | 14.5 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.3 | 4.8 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.3 | 13.4 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.3 | 3.0 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.3 | 1.2 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.3 | 13.2 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.3 | 1.4 | GO:0086045 | membrane depolarization during AV node cell action potential(GO:0086045) |
| 0.3 | 1.4 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.3 | 4.6 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.3 | 16.8 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.3 | 16.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.3 | 1.8 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) |
| 0.2 | 1.7 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.2 | 2.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.2 | 8.4 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.2 | 3.3 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.2 | 3.7 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.2 | 2.3 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.2 | 1.4 | GO:0042426 | choline catabolic process(GO:0042426) |
| 0.2 | 2.3 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.2 | 1.9 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.2 | 2.9 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.2 | 1.0 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.2 | 1.9 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.2 | 3.9 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.2 | 2.0 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.2 | 2.5 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.2 | 1.7 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.2 | 0.5 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.2 | 6.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.2 | 1.5 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.1 | 0.6 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 11.5 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 0.8 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 0.7 | GO:0035936 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.1 | 0.3 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.1 | 2.9 | GO:0051220 | cytoplasmic sequestering of protein(GO:0051220) |
| 0.1 | 3.4 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 2.0 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.1 | 1.6 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 0.5 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 4.2 | GO:0071173 | mitotic spindle assembly checkpoint(GO:0007094) spindle assembly checkpoint(GO:0071173) |
| 0.1 | 0.5 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.1 | 2.3 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 3.1 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 4.0 | GO:0009725 | response to hormone(GO:0009725) |
| 0.1 | 2.1 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 1.2 | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.1 | 0.4 | GO:0032900 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) negative regulation of neurotrophin production(GO:0032900) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 | 4.0 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.1 | 1.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 2.2 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.1 | 0.5 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 6.9 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 2.1 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.1 | 0.9 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 1.1 | GO:0042711 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.1 | 2.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.9 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 2.0 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.1 | 0.7 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 1.9 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.1 | 4.3 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 3.0 | GO:0048536 | spleen development(GO:0048536) |
| 0.1 | 0.1 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 14.9 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.1 | 4.5 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 25.1 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.1 | 1.5 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.1 | 3.2 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.1 | 4.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.4 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 6.1 | GO:1901216 | positive regulation of neuron death(GO:1901216) |
| 0.1 | 0.8 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.1 | 0.5 | GO:0015853 | adenine transport(GO:0015853) |
| 0.1 | 0.5 | GO:0007623 | circadian rhythm(GO:0007623) |
| 0.1 | 5.0 | GO:1904590 | negative regulation of protein import into nucleus(GO:0042308) negative regulation of protein import(GO:1904590) |
| 0.1 | 1.3 | GO:0007184 | SMAD protein import into nucleus(GO:0007184) |
| 0.1 | 0.7 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.1 | 3.8 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 4.0 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.1 | 2.1 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.1 | 8.6 | GO:0007626 | locomotory behavior(GO:0007626) |
| 0.1 | 1.4 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.1 | 1.0 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 3.5 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 1.2 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 1.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.9 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 2.6 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.1 | 3.8 | GO:0065002 | intracellular protein transmembrane transport(GO:0065002) |
| 0.1 | 4.5 | GO:0051301 | cell division(GO:0051301) |
| 0.1 | 0.5 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.1 | 0.4 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.7 | GO:0042753 | cell differentiation in hindbrain(GO:0021533) cerebellar Purkinje cell layer formation(GO:0021694) cerebellar cortex formation(GO:0021697) cerebellar Purkinje cell differentiation(GO:0021702) positive regulation of circadian rhythm(GO:0042753) |
| 0.1 | 6.7 | GO:0044070 | regulation of anion transport(GO:0044070) |
| 0.1 | 2.7 | GO:0006040 | amino sugar metabolic process(GO:0006040) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.2 | GO:1902613 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.0 | 0.1 | GO:0044065 | regulation of respiratory system process(GO:0044065) |
| 0.0 | 2.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 1.3 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.0 | 1.6 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 7.2 | GO:0010976 | positive regulation of neuron projection development(GO:0010976) |
| 0.0 | 0.2 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.0 | 22.8 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.3 | GO:0031175 | neuron projection development(GO:0031175) |
| 0.0 | 2.4 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.9 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 1.8 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 1.3 | GO:0097192 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.0 | 1.0 | GO:0009311 | oligosaccharide metabolic process(GO:0009311) |
| 0.0 | 0.6 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.2 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 2.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:1901160 | primary amino compound metabolic process(GO:1901160) |
| 0.0 | 0.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.2 | GO:0006388 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 1.2 | GO:0006641 | triglyceride metabolic process(GO:0006641) |
| 0.0 | 0.1 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 1.0 | GO:0045666 | positive regulation of neuron differentiation(GO:0045666) |
| 0.0 | 0.4 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.5 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 75.1 | GO:0005883 | neurofilament(GO:0005883) |
| 3.2 | 28.9 | GO:0005955 | calcineurin complex(GO:0005955) |
| 3.2 | 44.8 | GO:0097227 | sperm annulus(GO:0097227) |
| 2.8 | 2.8 | GO:0005683 | U7 snRNP(GO:0005683) |
| 2.2 | 41.3 | GO:0097386 | glial cell projection(GO:0097386) |
| 2.1 | 14.7 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 2.0 | 34.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 1.5 | 6.0 | GO:0032044 | DSIF complex(GO:0032044) |
| 1.4 | 2.9 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 1.4 | 11.1 | GO:0060203 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 1.4 | 8.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 1.3 | 22.7 | GO:0005687 | U4 snRNP(GO:0005687) |
| 1.2 | 36.2 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 1.1 | 5.7 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 1.1 | 22.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 1.1 | 12.4 | GO:0033270 | paranode region of axon(GO:0033270) |
| 1.1 | 10.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.8 | 10.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.6 | 5.5 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.6 | 49.8 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.6 | 57.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.6 | 1.7 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.6 | 3.4 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.6 | 3.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.5 | 10.9 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.5 | 3.8 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.5 | 13.0 | GO:0030673 | axolemma(GO:0030673) |
| 0.5 | 3.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.4 | 6.7 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.4 | 10.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.3 | 18.2 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.3 | 44.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.3 | 3.7 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.3 | 1.2 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.3 | 3.8 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.3 | 45.0 | GO:0000922 | spindle pole(GO:0000922) |
| 0.3 | 5.2 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.2 | 1.2 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.2 | 8.2 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.2 | 18.4 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.2 | 6.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 1.4 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.2 | 1.9 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.2 | 3.7 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 1.6 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.2 | 4.2 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.2 | 1.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.2 | 4.2 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.2 | 1.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 9.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 6.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 3.8 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.1 | 8.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 10.3 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 3.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 27.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 0.7 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 2.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 16.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 2.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 4.1 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.1 | 2.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 1.0 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 27.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 7.5 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 0.5 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 4.5 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 6.1 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 0.9 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 2.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 1.7 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 47.2 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.1 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.3 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 1.0 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 7.9 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.1 | 0.6 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.1 | 7.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.4 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 3.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 3.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.7 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 14.4 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.5 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 2.1 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.8 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 3.3 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 27.5 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 1.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.9 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.5 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 3.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 7.7 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 3.0 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 4.7 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 2.3 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 0.3 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 1.8 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0042611 | MHC protein complex(GO:0042611) MHC class II protein complex(GO:0042613) |
| 0.0 | 0.5 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 37.2 | 111.5 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 11.4 | 34.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 10.3 | 41.3 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 3.6 | 14.5 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 2.9 | 28.9 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 2.9 | 14.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 2.9 | 14.4 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 2.8 | 11.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 2.6 | 15.4 | GO:0044594 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 2.2 | 50.7 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 2.0 | 20.5 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 1.9 | 5.7 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 1.7 | 12.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 1.5 | 4.6 | GO:0019961 | interferon binding(GO:0019961) |
| 1.5 | 8.9 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 1.4 | 4.3 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 1.3 | 10.6 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 1.3 | 6.3 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 1.2 | 9.8 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 1.2 | 4.7 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 1.1 | 6.9 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 1.1 | 22.5 | GO:0008603 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 1.1 | 27.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 1.1 | 8.6 | GO:0004064 | arylesterase activity(GO:0004064) |
| 1.0 | 10.1 | GO:0004075 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 1.0 | 14.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 1.0 | 5.7 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.9 | 2.8 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.9 | 24.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.9 | 4.6 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.9 | 2.8 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.9 | 4.4 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.9 | 10.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.8 | 9.1 | GO:0051430 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.8 | 11.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.8 | 3.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.8 | 7.9 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.8 | 4.6 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.6 | 22.1 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.6 | 3.8 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.6 | 10.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.6 | 107.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.6 | 8.1 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.6 | 5.7 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.6 | 5.6 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.5 | 3.3 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.5 | 2.0 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.5 | 21.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.5 | 3.7 | GO:0019863 | immunoglobulin receptor activity(GO:0019763) IgE binding(GO:0019863) |
| 0.5 | 6.0 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.5 | 1.4 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.4 | 3.6 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.4 | 11.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.4 | 3.8 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.4 | 2.9 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.4 | 1.2 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.4 | 3.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.4 | 1.4 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.3 | 10.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.3 | 4.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.3 | 12.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.3 | 1.3 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.3 | 1.0 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.3 | 2.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.3 | 2.8 | GO:0043426 | MRF binding(GO:0043426) |
| 0.3 | 0.9 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.3 | 3.3 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.3 | 4.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.3 | 0.8 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.3 | 76.9 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.3 | 3.1 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.2 | 6.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.2 | 8.6 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.2 | 3.7 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.2 | 4.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 3.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.2 | 39.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.2 | 2.0 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.2 | 5.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 2.7 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 3.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.2 | 6.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.2 | 1.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 1.7 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.2 | 3.3 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.2 | 2.8 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.2 | 2.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.2 | 19.2 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.2 | 4.0 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.2 | 5.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 3.8 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.2 | 1.9 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 2.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.2 | 0.9 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.2 | 2.0 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 4.0 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 3.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 18.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 1.0 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 3.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 1.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 7.7 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 5.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 3.4 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 1.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 1.4 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 1.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.1 | 0.8 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 7.5 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 1.8 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 9.6 | GO:0020037 | heme binding(GO:0020037) |
| 0.1 | 0.2 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.1 | 2.9 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 3.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 4.0 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 17.3 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 4.6 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 0.5 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 1.0 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 0.7 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 0.6 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.1 | 0.7 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 3.5 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 4.3 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 7.0 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.1 | 0.3 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.1 | 1.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 3.1 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 3.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 7.4 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 12.3 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.9 | GO:0015278 | intracellular ligand-gated ion channel activity(GO:0005217) calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.2 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 1.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 2.1 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 2.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.9 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 1.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 10.1 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.2 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.6 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 3.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 4.6 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 1.7 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 1.1 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 3.7 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.5 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.5 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.1 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) |
| 0.0 | 1.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 111.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.8 | 52.0 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.6 | 10.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.6 | 28.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.6 | 3.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.4 | 12.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 18.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.3 | 16.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.3 | 19.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.3 | 4.6 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.3 | 3.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.3 | 15.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.3 | 20.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.3 | 6.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 1.7 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.2 | 5.6 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.2 | 4.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 3.5 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.2 | 3.4 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 3.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 2.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 1.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 21.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 3.7 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 10.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 2.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 2.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 3.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 5.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.2 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.8 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 2.9 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 2.2 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 1.1 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.6 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 111.5 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 3.2 | 67.4 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 2.4 | 23.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 1.3 | 35.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 1.2 | 15.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 1.0 | 35.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 1.0 | 50.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.8 | 3.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.8 | 5.7 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.8 | 21.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.7 | 10.2 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.6 | 8.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.6 | 10.6 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.6 | 19.6 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.6 | 15.6 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.6 | 11.6 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.4 | 4.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.4 | 11.5 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.4 | 4.5 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.3 | 15.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.3 | 8.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.3 | 6.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.3 | 7.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.3 | 3.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.3 | 3.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 4.9 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 4.0 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 4.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 3.8 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.2 | 8.1 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.2 | 2.7 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 10.4 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.2 | 3.1 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.2 | 7.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 7.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 1.2 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.1 | 2.9 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 2.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 1.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 3.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 4.5 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.1 | 7.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 16.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 3.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 2.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 8.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 2.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.6 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 4.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.4 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 1.7 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 2.7 | REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | Genes involved in Cleavage of Growing Transcript in the Termination Region |
| 0.0 | 1.9 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 4.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.3 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 1.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.6 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 1.1 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |