Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
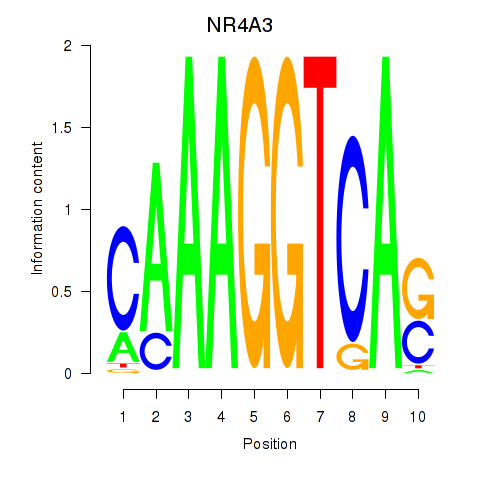
Results for NR4A3
Z-value: 0.45
Transcription factors associated with NR4A3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR4A3
|
ENSG00000119508.13 | nuclear receptor subfamily 4 group A member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR4A3 | hg19_v2_chr9_+_102584128_102584144 | -0.36 | 3.0e-08 | Click! |
Activity profile of NR4A3 motif
Sorted Z-values of NR4A3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_64059332 | 20.89 |
ENST00000540265.1
|
PGM1
|
phosphoglucomutase 1 |
| chr19_+_17416609 | 12.24 |
ENST00000602206.1
|
MRPL34
|
mitochondrial ribosomal protein L34 |
| chr10_+_81107271 | 11.02 |
ENST00000448165.1
|
PPIF
|
peptidylprolyl isomerase F |
| chr17_-_79995553 | 10.96 |
ENST00000581584.1
ENST00000577712.1 ENST00000582900.1 ENST00000579155.1 ENST00000306869.2 |
DCXR
|
dicarbonyl/L-xylulose reductase |
| chr17_-_4852332 | 9.95 |
ENST00000572383.1
|
PFN1
|
profilin 1 |
| chr5_+_1801503 | 9.29 |
ENST00000274137.5
ENST00000469176.1 |
NDUFS6
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6, 13kDa (NADH-coenzyme Q reductase) |
| chrX_-_53461288 | 9.21 |
ENST00000375298.4
ENST00000375304.5 |
HSD17B10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr20_+_35807449 | 9.18 |
ENST00000237530.6
|
RPN2
|
ribophorin II |
| chr19_+_17416457 | 9.00 |
ENST00000252602.1
|
MRPL34
|
mitochondrial ribosomal protein L34 |
| chr8_-_101718991 | 8.70 |
ENST00000517990.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr20_+_35807512 | 8.59 |
ENST00000373622.5
|
RPN2
|
ribophorin II |
| chr8_+_145149930 | 8.43 |
ENST00000318911.4
|
CYC1
|
cytochrome c-1 |
| chr5_-_95158644 | 8.21 |
ENST00000237858.6
|
GLRX
|
glutaredoxin (thioltransferase) |
| chr13_+_28194873 | 7.99 |
ENST00000302979.3
|
POLR1D
|
polymerase (RNA) I polypeptide D, 16kDa |
| chrX_-_53461305 | 7.78 |
ENST00000168216.6
|
HSD17B10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr11_+_35198243 | 7.69 |
ENST00000528455.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr2_-_209118974 | 7.65 |
ENST00000415913.1
ENST00000415282.1 ENST00000446179.1 |
IDH1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr1_-_40042416 | 7.56 |
ENST00000372857.3
ENST00000372856.3 ENST00000531243.2 ENST00000451091.2 |
PABPC4
|
poly(A) binding protein, cytoplasmic 4 (inducible form) |
| chr1_+_203830703 | 7.36 |
ENST00000414487.2
|
SNRPE
|
small nuclear ribonucleoprotein polypeptide E |
| chr11_-_64013663 | 7.29 |
ENST00000392210.2
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr11_-_73687997 | 6.68 |
ENST00000545212.1
|
UCP2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chrX_+_23801280 | 6.68 |
ENST00000379251.3
ENST00000379253.3 ENST00000379254.1 ENST00000379270.4 |
SAT1
|
spermidine/spermine N1-acetyltransferase 1 |
| chr5_+_68462837 | 6.59 |
ENST00000256442.5
|
CCNB1
|
cyclin B1 |
| chr6_-_43197189 | 6.39 |
ENST00000509253.1
ENST00000393987.2 ENST00000230431.6 |
DNPH1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr22_+_21128167 | 6.37 |
ENST00000215727.5
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr1_-_17380630 | 6.32 |
ENST00000375499.3
|
SDHB
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr11_+_35198118 | 6.11 |
ENST00000525211.1
ENST00000526000.1 ENST00000279452.6 ENST00000527889.1 |
CD44
|
CD44 molecule (Indian blood group) |
| chr7_+_128399002 | 6.09 |
ENST00000493278.1
|
CALU
|
calumenin |
| chr5_-_150466692 | 5.94 |
ENST00000315050.7
ENST00000523338.1 ENST00000522100.1 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr15_-_76603727 | 5.91 |
ENST00000560595.1
ENST00000433983.2 ENST00000559386.1 ENST00000559602.1 ENST00000560726.1 ENST00000557943.1 |
ETFA
|
electron-transfer-flavoprotein, alpha polypeptide |
| chr21_-_27107198 | 5.81 |
ENST00000400094.1
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr1_+_155179012 | 5.79 |
ENST00000609421.1
|
MTX1
|
metaxin 1 |
| chr20_-_60718430 | 5.63 |
ENST00000370873.4
ENST00000370858.3 |
PSMA7
|
proteasome (prosome, macropain) subunit, alpha type, 7 |
| chr22_+_20105259 | 5.59 |
ENST00000416427.1
ENST00000421656.1 ENST00000423859.1 ENST00000418705.2 |
RANBP1
|
RAN binding protein 1 |
| chr21_-_27107344 | 5.56 |
ENST00000457143.2
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr17_-_73178599 | 5.51 |
ENST00000578238.1
|
SUMO2
|
small ubiquitin-like modifier 2 |
| chr4_-_140216948 | 5.47 |
ENST00000265500.4
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr19_-_40336969 | 5.46 |
ENST00000599134.1
ENST00000597634.1 ENST00000598417.1 ENST00000601274.1 ENST00000594309.1 ENST00000221801.3 |
FBL
|
fibrillarin |
| chr15_-_65282274 | 5.30 |
ENST00000204566.2
|
SPG21
|
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chr1_+_53392901 | 5.29 |
ENST00000371514.3
ENST00000528311.1 ENST00000371509.4 ENST00000407246.2 ENST00000371513.5 |
SCP2
|
sterol carrier protein 2 |
| chr1_+_24019099 | 5.27 |
ENST00000443624.1
ENST00000458455.1 |
RPL11
|
ribosomal protein L11 |
| chr11_+_118272328 | 5.25 |
ENST00000524422.1
|
ATP5L
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G |
| chr11_-_62342375 | 5.21 |
ENST00000378019.3
|
EEF1G
|
eukaryotic translation elongation factor 1 gamma |
| chr3_+_120315160 | 5.19 |
ENST00000485064.1
ENST00000492739.1 |
NDUFB4
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4, 15kDa |
| chr15_-_65282232 | 5.18 |
ENST00000416889.2
|
SPG21
|
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chr6_-_31926629 | 5.17 |
ENST00000375425.5
ENST00000426722.1 ENST00000441998.1 ENST00000444811.2 ENST00000375429.3 |
NELFE
|
negative elongation factor complex member E |
| chr6_-_43027105 | 5.11 |
ENST00000230413.5
ENST00000487429.1 ENST00000489623.1 ENST00000468957.1 |
MRPL2
|
mitochondrial ribosomal protein L2 |
| chr8_-_80942139 | 5.07 |
ENST00000521434.1
ENST00000519120.1 ENST00000520946.1 |
MRPS28
|
mitochondrial ribosomal protein S28 |
| chrX_+_77359671 | 4.83 |
ENST00000373316.4
|
PGK1
|
phosphoglycerate kinase 1 |
| chr17_-_61973929 | 4.83 |
ENST00000329882.8
ENST00000453363.3 ENST00000316193.8 |
CSH1
|
chorionic somatomammotropin hormone 1 (placental lactogen) |
| chr17_-_61951090 | 4.80 |
ENST00000345366.7
ENST00000392886.2 ENST00000336844.5 ENST00000560142.1 |
CSH2
|
chorionic somatomammotropin hormone 2 |
| chr12_+_93861282 | 4.74 |
ENST00000552217.1
ENST00000393128.4 ENST00000547098.1 |
MRPL42
|
mitochondrial ribosomal protein L42 |
| chr1_-_155232221 | 4.72 |
ENST00000355379.3
|
SCAMP3
|
secretory carrier membrane protein 3 |
| chr2_+_88991162 | 4.66 |
ENST00000283646.4
|
RPIA
|
ribose 5-phosphate isomerase A |
| chrX_+_77359726 | 4.64 |
ENST00000442431.1
|
PGK1
|
phosphoglycerate kinase 1 |
| chr22_+_30163340 | 4.61 |
ENST00000330029.6
ENST00000401406.3 |
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr7_+_148892557 | 4.60 |
ENST00000262085.3
|
ZNF282
|
zinc finger protein 282 |
| chr17_-_73179046 | 4.55 |
ENST00000314523.7
ENST00000420826.2 |
SUMO2
|
small ubiquitin-like modifier 2 |
| chr14_-_23398565 | 4.53 |
ENST00000397440.4
ENST00000538452.1 ENST00000421938.2 ENST00000554867.1 ENST00000556616.1 ENST00000216350.8 ENST00000553550.1 ENST00000397441.2 ENST00000553897.1 |
PRMT5
|
protein arginine methyltransferase 5 |
| chr18_-_54318353 | 4.53 |
ENST00000590954.1
ENST00000540155.1 |
TXNL1
|
thioredoxin-like 1 |
| chr11_-_5271122 | 4.52 |
ENST00000330597.3
|
HBG1
|
hemoglobin, gamma A |
| chr21_-_27107283 | 4.52 |
ENST00000284971.3
ENST00000400099.1 |
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr11_+_67798363 | 4.50 |
ENST00000525628.1
|
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr1_-_155232047 | 4.47 |
ENST00000302631.3
|
SCAMP3
|
secretory carrier membrane protein 3 |
| chr11_+_34073195 | 4.44 |
ENST00000341394.4
|
CAPRIN1
|
cell cycle associated protein 1 |
| chr14_-_58893832 | 4.35 |
ENST00000556007.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr3_+_155588375 | 4.33 |
ENST00000295920.7
|
GMPS
|
guanine monphosphate synthase |
| chr2_+_98262497 | 4.31 |
ENST00000258424.2
|
COX5B
|
cytochrome c oxidase subunit Vb |
| chr14_-_105262055 | 4.29 |
ENST00000349310.3
|
AKT1
|
v-akt murine thymoma viral oncogene homolog 1 |
| chr11_+_67798114 | 4.24 |
ENST00000453471.2
ENST00000528492.1 ENST00000526339.1 ENST00000525419.1 |
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr1_-_33502528 | 4.22 |
ENST00000354858.6
|
AK2
|
adenylate kinase 2 |
| chr14_-_106209368 | 4.20 |
ENST00000390548.2
ENST00000390549.2 ENST00000390542.2 |
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker) |
| chr3_+_179322573 | 4.19 |
ENST00000493866.1
ENST00000472629.1 ENST00000482604.1 |
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr19_+_47634039 | 4.19 |
ENST00000597808.1
ENST00000413379.3 ENST00000600706.1 ENST00000540850.1 ENST00000598840.1 ENST00000600753.1 ENST00000270225.7 ENST00000392776.3 |
SAE1
|
SUMO1 activating enzyme subunit 1 |
| chr9_-_130637244 | 4.17 |
ENST00000373156.1
|
AK1
|
adenylate kinase 1 |
| chr16_+_2255841 | 4.16 |
ENST00000301725.7
|
MLST8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr20_-_57617831 | 4.14 |
ENST00000371033.5
ENST00000355937.4 |
SLMO2
|
slowmo homolog 2 (Drosophila) |
| chr1_+_46769303 | 4.12 |
ENST00000311672.5
|
UQCRH
|
ubiquinol-cytochrome c reductase hinge protein |
| chr7_+_142960505 | 4.12 |
ENST00000409500.3
ENST00000443571.2 ENST00000358406.5 ENST00000479303.1 |
GSTK1
|
glutathione S-transferase kappa 1 |
| chr18_+_29077990 | 4.07 |
ENST00000261590.8
|
DSG2
|
desmoglein 2 |
| chr12_+_98987369 | 3.98 |
ENST00000401722.3
ENST00000188376.5 ENST00000228318.3 ENST00000551917.1 ENST00000548046.1 ENST00000552981.1 ENST00000551265.1 ENST00000550695.1 ENST00000547534.1 ENST00000549338.1 ENST00000548847.1 |
SLC25A3
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3 |
| chr10_+_81272287 | 3.97 |
ENST00000520547.2
|
EIF5AL1
|
eukaryotic translation initiation factor 5A-like 1 |
| chr17_+_4854375 | 3.96 |
ENST00000521811.1
ENST00000519602.1 ENST00000323997.6 ENST00000522249.1 ENST00000519584.1 |
ENO3
|
enolase 3 (beta, muscle) |
| chr18_+_55816546 | 3.95 |
ENST00000435432.2
ENST00000357895.5 ENST00000586263.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr16_+_88872176 | 3.94 |
ENST00000569140.1
|
CDT1
|
chromatin licensing and DNA replication factor 1 |
| chr16_+_2255710 | 3.94 |
ENST00000397124.1
ENST00000565250.1 |
MLST8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr12_-_56709674 | 3.92 |
ENST00000551286.1
ENST00000549318.1 |
CNPY2
RP11-977G19.10
|
canopy FGF signaling regulator 2 Uncharacterized protein |
| chr6_+_31633902 | 3.90 |
ENST00000375865.2
ENST00000375866.2 |
CSNK2B
|
casein kinase 2, beta polypeptide |
| chr5_+_218356 | 3.90 |
ENST00000264932.6
ENST00000504309.1 ENST00000510361.1 |
SDHA
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp) |
| chr1_+_45205498 | 3.89 |
ENST00000372218.4
|
KIF2C
|
kinesin family member 2C |
| chr1_-_229569834 | 3.88 |
ENST00000366684.3
ENST00000366683.2 |
ACTA1
|
actin, alpha 1, skeletal muscle |
| chr11_-_66206260 | 3.88 |
ENST00000329819.4
ENST00000310999.7 ENST00000430466.2 |
MRPL11
|
mitochondrial ribosomal protein L11 |
| chr22_+_31518938 | 3.87 |
ENST00000412985.1
ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr22_+_38609538 | 3.84 |
ENST00000407965.1
|
MAFF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr18_+_3412005 | 3.82 |
ENST00000401449.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr7_-_140179276 | 3.77 |
ENST00000443720.2
ENST00000255977.2 |
MKRN1
|
makorin ring finger protein 1 |
| chr8_-_101734170 | 3.76 |
ENST00000522387.1
ENST00000518196.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr1_+_45205478 | 3.71 |
ENST00000452259.1
ENST00000372224.4 |
KIF2C
|
kinesin family member 2C |
| chr6_-_41909466 | 3.67 |
ENST00000414200.2
|
CCND3
|
cyclin D3 |
| chr14_-_24610779 | 3.66 |
ENST00000560403.1
ENST00000419198.2 ENST00000216799.4 |
EMC9
|
ER membrane protein complex subunit 9 |
| chr11_+_34073757 | 3.62 |
ENST00000532820.1
|
CAPRIN1
|
cell cycle associated protein 1 |
| chr22_+_31488433 | 3.59 |
ENST00000455608.1
|
SMTN
|
smoothelin |
| chr20_+_34129770 | 3.59 |
ENST00000348547.2
ENST00000357394.4 ENST00000447986.1 ENST00000279052.6 ENST00000416206.1 ENST00000411577.1 ENST00000413587.1 |
ERGIC3
|
ERGIC and golgi 3 |
| chr17_-_64216748 | 3.56 |
ENST00000585162.1
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr13_+_25670268 | 3.53 |
ENST00000281589.3
|
PABPC3
|
poly(A) binding protein, cytoplasmic 3 |
| chr14_-_104387831 | 3.50 |
ENST00000557040.1
ENST00000414262.2 ENST00000555030.1 ENST00000554713.1 ENST00000553430.1 |
C14orf2
|
chromosome 14 open reading frame 2 |
| chr11_-_61560254 | 3.50 |
ENST00000543510.1
|
TMEM258
|
transmembrane protein 258 |
| chr6_-_41909561 | 3.49 |
ENST00000372991.4
|
CCND3
|
cyclin D3 |
| chr14_-_104387888 | 3.46 |
ENST00000286953.3
|
C14orf2
|
chromosome 14 open reading frame 2 |
| chr12_+_104359576 | 3.46 |
ENST00000392872.3
ENST00000436021.2 |
TDG
|
thymine-DNA glycosylase |
| chr16_-_67969888 | 3.41 |
ENST00000574576.2
|
PSMB10
|
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr17_+_4855053 | 3.39 |
ENST00000518175.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr18_+_11981547 | 3.38 |
ENST00000588927.1
|
IMPA2
|
inositol(myo)-1(or 4)-monophosphatase 2 |
| chr1_-_1711508 | 3.38 |
ENST00000378625.1
|
NADK
|
NAD kinase |
| chr19_-_2427536 | 3.38 |
ENST00000591871.1
|
TIMM13
|
translocase of inner mitochondrial membrane 13 homolog (yeast) |
| chr11_+_65769946 | 3.33 |
ENST00000533166.1
|
BANF1
|
barrier to autointegration factor 1 |
| chr13_-_31038370 | 3.32 |
ENST00000399489.1
ENST00000339872.4 |
HMGB1
|
high mobility group box 1 |
| chr10_+_22605304 | 3.29 |
ENST00000475460.2
ENST00000602390.1 ENST00000489125.2 ENST00000456711.1 ENST00000444869.1 |
COMMD3-BMI1
COMMD3
|
COMMD3-BMI1 readthrough COMM domain containing 3 |
| chr1_+_182992545 | 3.29 |
ENST00000258341.4
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2) |
| chr2_+_109271481 | 3.28 |
ENST00000542845.1
ENST00000393314.2 |
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr11_+_125496619 | 3.24 |
ENST00000532669.1
ENST00000278916.3 |
CHEK1
|
checkpoint kinase 1 |
| chr2_+_220363579 | 3.16 |
ENST00000313597.5
ENST00000373917.3 ENST00000358215.3 ENST00000373908.1 ENST00000455657.1 ENST00000435316.1 ENST00000341142.3 |
GMPPA
|
GDP-mannose pyrophosphorylase A |
| chr5_-_39425068 | 3.15 |
ENST00000515700.1
ENST00000339788.6 |
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr1_-_173886491 | 3.14 |
ENST00000367698.3
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1 |
| chr15_+_75335604 | 3.12 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr18_+_55888767 | 3.10 |
ENST00000431212.2
ENST00000586268.1 ENST00000587190.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr1_+_151129135 | 3.09 |
ENST00000602841.1
|
SCNM1
|
sodium channel modifier 1 |
| chr12_-_122296755 | 3.07 |
ENST00000289004.4
|
HPD
|
4-hydroxyphenylpyruvate dioxygenase |
| chr11_+_125496400 | 3.07 |
ENST00000524737.1
|
CHEK1
|
checkpoint kinase 1 |
| chr16_+_4674787 | 3.06 |
ENST00000262370.7
|
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr8_-_101734308 | 3.04 |
ENST00000519004.1
ENST00000519363.1 ENST00000520142.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr20_-_17641097 | 3.03 |
ENST00000246043.4
|
RRBP1
|
ribosome binding protein 1 |
| chr14_+_23791159 | 3.00 |
ENST00000557702.1
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
| chr2_-_37458749 | 2.98 |
ENST00000234170.5
|
CEBPZ
|
CCAAT/enhancer binding protein (C/EBP), zeta |
| chr8_-_131028660 | 2.98 |
ENST00000401979.2
ENST00000517654.1 ENST00000522361.1 ENST00000518167.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr1_-_33502441 | 2.97 |
ENST00000548033.1
ENST00000487289.1 ENST00000373449.2 ENST00000480134.1 ENST00000467905.1 |
AK2
|
adenylate kinase 2 |
| chr12_+_120875910 | 2.96 |
ENST00000551806.1
|
AL021546.6
|
Glutamyl-tRNA(Gln) amidotransferase subunit C, mitochondrial |
| chr2_+_26467825 | 2.95 |
ENST00000545822.1
ENST00000425035.1 |
HADHB
|
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), beta subunit |
| chr14_+_95047744 | 2.94 |
ENST00000553511.1
ENST00000554633.1 ENST00000555681.1 ENST00000554276.1 |
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr14_-_105262016 | 2.93 |
ENST00000407796.2
|
AKT1
|
v-akt murine thymoma viral oncogene homolog 1 |
| chr11_-_61560053 | 2.91 |
ENST00000537328.1
|
TMEM258
|
transmembrane protein 258 |
| chr8_-_145018905 | 2.90 |
ENST00000398774.2
|
PLEC
|
plectin |
| chr1_-_33815486 | 2.89 |
ENST00000373418.3
|
PHC2
|
polyhomeotic homolog 2 (Drosophila) |
| chr8_-_144897138 | 2.88 |
ENST00000377533.3
|
SCRIB
|
scribbled planar cell polarity protein |
| chr3_+_160117418 | 2.86 |
ENST00000465903.1
ENST00000485645.1 ENST00000360111.2 ENST00000472991.1 ENST00000467468.1 ENST00000469762.1 ENST00000489573.1 ENST00000462787.1 ENST00000490207.1 ENST00000485867.1 |
SMC4
|
structural maintenance of chromosomes 4 |
| chr7_-_100239132 | 2.85 |
ENST00000223051.3
ENST00000431692.1 |
TFR2
|
transferrin receptor 2 |
| chr22_+_38245414 | 2.77 |
ENST00000381683.6
ENST00000414316.1 ENST00000406934.1 ENST00000451427.1 |
EIF3L
|
eukaryotic translation initiation factor 3, subunit L |
| chr16_+_4674814 | 2.77 |
ENST00000415496.1
ENST00000587747.1 ENST00000399577.5 ENST00000588994.1 ENST00000586183.1 |
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr1_-_29508499 | 2.77 |
ENST00000373795.4
|
SRSF4
|
serine/arginine-rich splicing factor 4 |
| chr20_+_43211149 | 2.76 |
ENST00000372886.1
|
PKIG
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr11_+_66824346 | 2.75 |
ENST00000532559.1
|
RHOD
|
ras homolog family member D |
| chr14_+_74035763 | 2.74 |
ENST00000238651.5
|
ACOT2
|
acyl-CoA thioesterase 2 |
| chr1_-_120311517 | 2.74 |
ENST00000369406.3
ENST00000544913.2 |
HMGCS2
|
3-hydroxy-3-methylglutaryl-CoA synthase 2 (mitochondrial) |
| chr12_-_6451186 | 2.73 |
ENST00000540022.1
ENST00000536194.1 |
TNFRSF1A
|
tumor necrosis factor receptor superfamily, member 1A |
| chr7_+_6048856 | 2.72 |
ENST00000223029.3
ENST00000400479.2 ENST00000395236.2 |
AIMP2
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 |
| chr14_+_35452104 | 2.71 |
ENST00000216774.6
ENST00000546080.1 |
SRP54
|
signal recognition particle 54kDa |
| chr16_-_47007545 | 2.71 |
ENST00000317089.5
|
DNAJA2
|
DnaJ (Hsp40) homolog, subfamily A, member 2 |
| chr3_+_122785895 | 2.68 |
ENST00000316218.7
|
PDIA5
|
protein disulfide isomerase family A, member 5 |
| chr3_+_25831567 | 2.67 |
ENST00000280701.3
ENST00000420173.2 |
OXSM
|
3-oxoacyl-ACP synthase, mitochondrial |
| chr20_+_57466357 | 2.66 |
ENST00000371095.3
ENST00000371085.3 ENST00000354359.7 ENST00000265620.7 |
GNAS
|
GNAS complex locus |
| chr19_+_41117770 | 2.66 |
ENST00000601032.1
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr2_-_238499303 | 2.64 |
ENST00000409576.1
|
RAB17
|
RAB17, member RAS oncogene family |
| chr12_+_117013656 | 2.64 |
ENST00000556529.1
|
MAP1LC3B2
|
microtubule-associated protein 1 light chain 3 beta 2 |
| chr6_+_32938665 | 2.63 |
ENST00000374831.4
ENST00000395289.2 |
BRD2
|
bromodomain containing 2 |
| chr19_+_19303008 | 2.63 |
ENST00000353145.1
ENST00000421262.3 ENST00000303088.4 ENST00000456252.3 ENST00000593273.1 |
RFXANK
|
regulatory factor X-associated ankyrin-containing protein |
| chr16_+_81040103 | 2.62 |
ENST00000305850.5
ENST00000299572.5 |
CENPN
|
centromere protein N |
| chr3_-_129158850 | 2.61 |
ENST00000503197.1
ENST00000249910.1 ENST00000429544.2 ENST00000507208.1 |
MBD4
|
methyl-CpG binding domain protein 4 |
| chr1_-_27701307 | 2.59 |
ENST00000270879.4
ENST00000354982.2 |
FCN3
|
ficolin (collagen/fibrinogen domain containing) 3 |
| chr15_+_58724184 | 2.58 |
ENST00000433326.2
|
LIPC
|
lipase, hepatic |
| chr11_+_66824276 | 2.57 |
ENST00000308831.2
|
RHOD
|
ras homolog family member D |
| chr16_+_72090053 | 2.57 |
ENST00000576168.2
ENST00000567185.3 ENST00000567612.2 |
HP
|
haptoglobin |
| chr19_+_42363917 | 2.56 |
ENST00000598742.1
|
RPS19
|
ribosomal protein S19 |
| chr16_-_30538079 | 2.55 |
ENST00000562803.1
|
ZNF768
|
zinc finger protein 768 |
| chrX_-_77225135 | 2.54 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr1_-_17304771 | 2.52 |
ENST00000375534.3
|
MFAP2
|
microfibrillar-associated protein 2 |
| chr6_-_100016678 | 2.50 |
ENST00000523799.1
ENST00000520429.1 |
CCNC
|
cyclin C |
| chr1_+_150480576 | 2.50 |
ENST00000346569.6
|
ECM1
|
extracellular matrix protein 1 |
| chr1_-_165738072 | 2.49 |
ENST00000481278.1
|
TMCO1
|
transmembrane and coiled-coil domains 1 |
| chr14_-_106111127 | 2.49 |
ENST00000390545.2
|
IGHG2
|
immunoglobulin heavy constant gamma 2 (G2m marker) |
| chr5_+_179233376 | 2.48 |
ENST00000376929.3
ENST00000514093.1 |
SQSTM1
|
sequestosome 1 |
| chr17_-_17494972 | 2.48 |
ENST00000435340.2
ENST00000255389.5 ENST00000395781.2 |
PEMT
|
phosphatidylethanolamine N-methyltransferase |
| chr17_+_55173933 | 2.45 |
ENST00000539273.1
|
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr5_+_140019004 | 2.44 |
ENST00000394671.3
ENST00000511410.1 ENST00000537378.1 |
TMCO6
|
transmembrane and coiled-coil domains 6 |
| chr7_-_16840820 | 2.40 |
ENST00000450569.1
|
AGR2
|
anterior gradient 2 |
| chr1_+_151138500 | 2.40 |
ENST00000368905.4
|
SCNM1
|
sodium channel modifier 1 |
| chr19_-_49552363 | 2.37 |
ENST00000448456.3
ENST00000355414.2 |
CGB8
|
chorionic gonadotropin, beta polypeptide 8 |
| chrX_-_134049262 | 2.35 |
ENST00000370783.3
|
MOSPD1
|
motile sperm domain containing 1 |
| chr11_+_7618413 | 2.34 |
ENST00000528883.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr9_+_115983808 | 2.32 |
ENST00000374210.6
ENST00000374212.4 |
SLC31A1
|
solute carrier family 31 (copper transporter), member 1 |
| chr7_-_139763521 | 2.31 |
ENST00000263549.3
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr10_+_94050913 | 2.31 |
ENST00000358935.2
|
MARCH5
|
membrane-associated ring finger (C3HC4) 5 |
| chr19_-_15236470 | 2.29 |
ENST00000533747.1
ENST00000598709.1 ENST00000534378.1 |
ILVBL
|
ilvB (bacterial acetolactate synthase)-like |
| chr2_+_68592305 | 2.29 |
ENST00000234313.7
|
PLEK
|
pleckstrin |
| chr9_-_75567962 | 2.23 |
ENST00000297785.3
ENST00000376939.1 |
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr1_-_46769261 | 2.22 |
ENST00000343304.6
|
LRRC41
|
leucine rich repeat containing 41 |
| chr3_+_186435137 | 2.21 |
ENST00000447445.1
|
KNG1
|
kininogen 1 |
| chr4_-_40517984 | 2.21 |
ENST00000381795.6
|
RBM47
|
RNA binding motif protein 47 |
| chr5_-_68665815 | 2.20 |
ENST00000380818.3
ENST00000328663.4 |
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chr17_-_77924627 | 2.19 |
ENST00000572862.1
ENST00000573782.1 ENST00000574427.1 ENST00000570373.1 ENST00000340848.7 ENST00000576768.1 |
TBC1D16
|
TBC1 domain family, member 16 |
| chr16_+_31539183 | 2.17 |
ENST00000302312.4
|
AHSP
|
alpha hemoglobin stabilizing protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR4A3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 17.0 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 3.7 | 11.0 | GO:2000276 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 2.7 | 11.0 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 2.6 | 7.7 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 2.4 | 7.2 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 2.2 | 6.7 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 2.2 | 6.6 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to DDT(GO:0046680) histone H3-S10 phosphorylation involved in chromosome condensation(GO:2000775) |
| 2.0 | 5.9 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 1.8 | 7.2 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 1.7 | 13.8 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 1.6 | 20.9 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 1.6 | 4.8 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 1.5 | 13.4 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 1.4 | 7.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 1.4 | 6.9 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 1.4 | 5.5 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 1.3 | 5.3 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 1.3 | 3.9 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 1.3 | 3.9 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 1.2 | 17.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 1.2 | 4.7 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 1.1 | 4.2 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 1.1 | 5.3 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 1.1 | 6.3 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 1.0 | 3.0 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 1.0 | 3.0 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 1.0 | 10.0 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 1.0 | 2.9 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.9 | 6.4 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.9 | 3.5 | GO:1902544 | oxidative DNA demethylation(GO:0035511) regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.9 | 7.7 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.9 | 2.6 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.9 | 26.4 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.8 | 3.3 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) regulation of restriction endodeoxyribonuclease activity(GO:0032072) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.8 | 9.0 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.8 | 4.1 | GO:0003164 | His-Purkinje system development(GO:0003164) |
| 0.8 | 4.8 | GO:0035709 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.8 | 3.2 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.8 | 7.6 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.7 | 2.2 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.7 | 4.3 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.7 | 5.5 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.7 | 3.4 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.7 | 6.7 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.6 | 3.9 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.6 | 2.6 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.6 | 1.9 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.6 | 2.5 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.6 | 6.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.6 | 3.0 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.6 | 3.0 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.6 | 1.8 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.6 | 42.4 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.6 | 1.7 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.6 | 1.7 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.6 | 11.3 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.6 | 3.4 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.6 | 1.1 | GO:0019860 | uracil metabolic process(GO:0019860) |
| 0.5 | 7.1 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.5 | 4.9 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.5 | 36.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.5 | 1.6 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.5 | 2.7 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.5 | 8.2 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.5 | 1.5 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.5 | 3.0 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.5 | 3.9 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.5 | 1.0 | GO:0003285 | septum secundum development(GO:0003285) |
| 0.5 | 5.8 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.5 | 2.8 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.5 | 1.8 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.4 | 3.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.4 | 4.8 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.4 | 3.9 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.4 | 1.7 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) phenylpropanoid catabolic process(GO:0046271) |
| 0.4 | 0.4 | GO:0050787 | detoxification of mercury ion(GO:0050787) |
| 0.4 | 2.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.4 | 2.0 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.4 | 4.0 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.4 | 2.4 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.4 | 5.5 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.4 | 6.2 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.4 | 2.3 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.4 | 2.3 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.4 | 2.7 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.4 | 2.3 | GO:0051643 | regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031585) secretory granule localization(GO:0032252) endoplasmic reticulum localization(GO:0051643) |
| 0.4 | 0.7 | GO:0071423 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.4 | 2.6 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.4 | 4.4 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.4 | 2.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.4 | 2.2 | GO:1901098 | regulation of autophagosome maturation(GO:1901096) positive regulation of autophagosome maturation(GO:1901098) |
| 0.4 | 2.9 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.3 | 2.8 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.3 | 1.0 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.3 | 1.7 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.3 | 2.0 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.3 | 1.3 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.3 | 1.3 | GO:0014040 | positive regulation of Schwann cell differentiation(GO:0014040) |
| 0.3 | 2.5 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.3 | 18.4 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.3 | 1.9 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.3 | 2.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.3 | 0.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.3 | 1.5 | GO:1902513 | regulation of organelle transport along microtubule(GO:1902513) |
| 0.3 | 4.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.3 | 1.2 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.3 | 3.0 | GO:0006983 | ER overload response(GO:0006983) |
| 0.3 | 1.5 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.3 | 6.4 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.3 | 1.7 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.3 | 3.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.3 | 1.4 | GO:0097021 | Peyer's patch morphogenesis(GO:0061146) lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.3 | 1.7 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.3 | 8.7 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.3 | 8.1 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.3 | 4.6 | GO:0042775 | ATP synthesis coupled electron transport(GO:0042773) mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.3 | 2.6 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.3 | 7.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.3 | 1.8 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.3 | 0.8 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.3 | 2.1 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.3 | 1.8 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.2 | 0.7 | GO:0050894 | determination of affect(GO:0050894) |
| 0.2 | 3.4 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.2 | 1.2 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.2 | 3.9 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.2 | 1.4 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.2 | 1.2 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.2 | 1.6 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.2 | 1.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.2 | 0.7 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.2 | 0.9 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.2 | 4.0 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.2 | 1.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.2 | 12.3 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.2 | 9.1 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.2 | 4.8 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.2 | 2.5 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.2 | 4.4 | GO:0007567 | parturition(GO:0007567) |
| 0.2 | 6.8 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.2 | 2.2 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.2 | 0.6 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.2 | 3.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.2 | 7.9 | GO:0061621 | NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.2 | 0.2 | GO:1990641 | response to iron ion starvation(GO:1990641) |
| 0.2 | 0.4 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.2 | 1.7 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.2 | 0.6 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.2 | 0.6 | GO:0070781 | arginine biosynthetic process via ornithine(GO:0042450) response to biotin(GO:0070781) |
| 0.2 | 2.0 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.2 | 8.0 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.2 | 2.8 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 2.7 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.2 | 1.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.2 | 0.5 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.2 | 1.0 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.2 | 1.5 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.2 | 2.3 | GO:0090344 | negative regulation of cell aging(GO:0090344) |
| 0.2 | 2.3 | GO:0034370 | triglyceride-rich lipoprotein particle remodeling(GO:0034370) very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.2 | 0.9 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.2 | 1.8 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.2 | 4.8 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.1 | 1.2 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.1 | 0.4 | GO:1903598 | positive regulation of gap junction assembly(GO:1903598) |
| 0.1 | 1.5 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.1 | 0.8 | GO:0030421 | defecation(GO:0030421) |
| 0.1 | 0.8 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 2.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.5 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.1 | 2.3 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 1.9 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.1 | 1.5 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.4 | GO:0051796 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.1 | 0.4 | GO:1903121 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.1 | 2.0 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 1.1 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.1 | 4.2 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.1 | 1.9 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 0.1 | 0.6 | GO:0045963 | cellular response to phosphate starvation(GO:0016036) positive regulation of sulfur amino acid metabolic process(GO:0031337) negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.1 | 1.2 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.1 | 0.7 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.1 | 9.8 | GO:0090109 | regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.1 | 1.0 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.1 | 0.6 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 9.4 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.1 | 7.2 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.1 | 0.8 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 0.3 | GO:0060127 | subthalamus development(GO:0021539) subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.1 | 1.9 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) negative regulation of biomineral tissue development(GO:0070168) |
| 0.1 | 2.8 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 5.1 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 2.6 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 0.6 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 1.4 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 1.9 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 6.1 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 0.7 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.1 | 0.7 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.1 | 1.5 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.1 | 2.7 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 3.0 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.1 | 0.5 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.1 | 2.7 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 0.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.7 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.1 | 1.7 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.1 | 1.2 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) |
| 0.1 | 1.3 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.1 | 2.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.1 | GO:0002442 | serotonin production involved in inflammatory response(GO:0002351) serotonin secretion involved in inflammatory response(GO:0002442) serotonin secretion by platelet(GO:0002554) |
| 0.1 | 1.8 | GO:0016071 | mRNA metabolic process(GO:0016071) |
| 0.1 | 1.7 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.1 | 1.5 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.1 | 1.0 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.1 | 1.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 6.8 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.1 | 0.5 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 1.5 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 0.4 | GO:0006701 | progesterone biosynthetic process(GO:0006701) Sertoli cell proliferation(GO:0060011) |
| 0.1 | 0.7 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.1 | 0.6 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.1 | 1.3 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.1 | 1.5 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.1 | 0.3 | GO:1902336 | vestibulocochlear nerve structural organization(GO:0021649) neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) positive regulation of cytokine activity(GO:0060301) renal artery morphogenesis(GO:0061441) ganglion morphogenesis(GO:0061552) endothelial tip cell fate specification(GO:0097102) positive regulation of retinal ganglion cell axon guidance(GO:1902336) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.1 | 2.2 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.0 | 0.1 | GO:0032006 | regulation of TOR signaling(GO:0032006) |
| 0.0 | 1.3 | GO:0070873 | regulation of glycogen metabolic process(GO:0070873) |
| 0.0 | 0.8 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 6.7 | GO:0038096 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.0 | 0.2 | GO:0060694 | regulation of cholesterol transporter activity(GO:0060694) response to metformin(GO:1901558) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 5.5 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.0 | 0.1 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.0 | 0.6 | GO:0007588 | excretion(GO:0007588) |
| 0.0 | 1.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.9 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.1 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.0 | 1.7 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 3.8 | GO:0002504 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 | 1.2 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.5 | GO:1901798 | positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 2.6 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.7 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.6 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.4 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.0 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 2.1 | GO:0035113 | embryonic limb morphogenesis(GO:0030326) embryonic appendage morphogenesis(GO:0035113) |
| 0.0 | 4.9 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.0 | 1.0 | GO:1903363 | negative regulation of cellular protein catabolic process(GO:1903363) |
| 0.0 | 1.6 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.2 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 2.0 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.0 | 0.2 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 17.0 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 2.6 | 10.2 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 2.0 | 24.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 2.0 | 13.8 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 1.8 | 5.5 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 1.6 | 17.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 1.3 | 6.6 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 1.2 | 7.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 1.2 | 21.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 1.1 | 4.3 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 1.0 | 4.2 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 1.0 | 5.2 | GO:0032021 | NELF complex(GO:0032021) |
| 1.0 | 8.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 1.0 | 2.9 | GO:0036026 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.9 | 37.1 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.9 | 7.4 | GO:0034715 | U7 snRNP(GO:0005683) pICln-Sm protein complex(GO:0034715) |
| 0.9 | 12.0 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.8 | 3.3 | GO:0043260 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 0.8 | 15.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.7 | 8.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.7 | 3.4 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.6 | 4.5 | GO:0034709 | methylosome(GO:0034709) |
| 0.6 | 2.6 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.6 | 4.8 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.6 | 2.9 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.6 | 5.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.6 | 36.5 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.6 | 3.9 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.5 | 6.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.5 | 5.6 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.5 | 7.7 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.5 | 1.4 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.5 | 1.4 | GO:0071749 | IgA immunoglobulin complex(GO:0071745) IgA immunoglobulin complex, circulating(GO:0071746) monomeric IgA immunoglobulin complex(GO:0071748) polymeric IgA immunoglobulin complex(GO:0071749) secretory IgA immunoglobulin complex(GO:0071751) |
| 0.4 | 8.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.4 | 3.4 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.4 | 1.3 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.4 | 1.2 | GO:0097679 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) other organism cytoplasm(GO:0097679) |
| 0.4 | 4.0 | GO:0038201 | TORC2 complex(GO:0031932) TOR complex(GO:0038201) |
| 0.4 | 30.5 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.4 | 3.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.3 | 2.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.3 | 10.5 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.3 | 17.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.3 | 2.9 | GO:0000796 | condensin complex(GO:0000796) |
| 0.3 | 2.2 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.2 | 2.0 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.2 | 3.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 2.8 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.2 | 4.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.2 | 1.3 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 2.5 | GO:0044754 | autolysosome(GO:0044754) |
| 0.2 | 2.9 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.2 | 5.1 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.2 | 11.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 3.0 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.2 | 13.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 1.5 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.2 | 0.7 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.2 | 1.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 3.0 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 1.5 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 1.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 4.8 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.1 | 2.7 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 2.9 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.4 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 2.8 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 5.1 | GO:0098576 | lumenal side of membrane(GO:0098576) |
| 0.1 | 14.4 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 1.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 5.0 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 3.6 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.1 | 1.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.7 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 1.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 4.5 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 0.9 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 11.9 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 2.0 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 4.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 3.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 5.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 3.9 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 5.9 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.1 | 2.1 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 3.7 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 6.1 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 4.9 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.1 | 3.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 19.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 1.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 12.6 | GO:0005840 | ribosome(GO:0005840) |
| 0.1 | 4.4 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 4.6 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 2.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 2.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 14.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 2.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.3 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 2.2 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 1.9 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.1 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 2.3 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 1.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.5 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 9.0 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.4 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.7 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 3.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.7 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.8 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.2 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.3 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.4 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 1.5 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 20.9 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 3.4 | 10.2 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 3.2 | 9.5 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 2.8 | 8.4 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 2.6 | 7.7 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 2.5 | 7.6 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 1.9 | 5.6 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 1.8 | 17.8 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 1.8 | 5.3 | GO:0070538 | oleic acid binding(GO:0070538) |
| 1.3 | 8.8 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 1.2 | 7.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 1.2 | 6.1 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 1.1 | 4.2 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 1.1 | 6.3 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 1.0 | 19.9 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 1.0 | 4.8 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 1.0 | 4.8 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.9 | 11.4 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.9 | 6.6 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.9 | 15.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.9 | 4.5 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.9 | 7.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.8 | 10.1 | GO:0031386 | protein tag(GO:0031386) |
| 0.8 | 10.0 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.8 | 2.5 | GO:0080101 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.8 | 8.2 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.8 | 4.8 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.8 | 11.0 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.8 | 7.6 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.7 | 4.5 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.7 | 7.4 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.7 | 2.8 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.7 | 3.6 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.7 | 2.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.7 | 4.2 | GO:0044388 | ubiquitin activating enzyme activity(GO:0004839) small protein activating enzyme binding(GO:0044388) |
| 0.7 | 3.4 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.7 | 2.0 | GO:0002134 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.7 | 2.0 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.7 | 34.8 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.6 | 1.9 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.6 | 2.5 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.6 | 11.0 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.6 | 0.6 | GO:0016679 | oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) |
| 0.6 | 1.7 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.6 | 1.7 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.6 | 3.3 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.5 | 2.7 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.5 | 5.3 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.5 | 10.5 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.5 | 1.9 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.5 | 13.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.5 | 1.8 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.4 | 2.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.4 | 8.0 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.4 | 1.7 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.4 | 7.2 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.4 | 4.7 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.4 | 1.7 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.4 | 2.5 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.4 | 4.5 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.4 | 3.9 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.4 | 1.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.4 | 5.5 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.3 | 2.9 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.3 | 4.3 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.3 | 1.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.3 | 9.0 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.3 | 2.7 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.3 | 0.9 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.3 | 2.0 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.3 | 1.7 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.3 | 2.7 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.3 | 7.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 0.7 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.2 | 8.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.2 | 2.7 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.2 | 2.7 | GO:0051430 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.2 | 6.4 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.2 | 2.3 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.2 | 26.1 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.2 | 2.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.2 | 1.6 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.2 | 1.8 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.2 | 0.4 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.2 | 4.0 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.2 | 2.8 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.2 | 9.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.2 | 5.5 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.2 | 1.5 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 1.8 | GO:0030883 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.2 | 3.9 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.2 | 0.9 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.2 | 37.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.2 | 3.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.2 | 4.8 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.2 | 4.8 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 2.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.2 | 7.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.2 | 2.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.2 | 1.2 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 1.5 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.2 | 1.7 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 5.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.6 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 1.0 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.6 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.1 | 3.2 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.6 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 6.5 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 0.6 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 12.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 1.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 5.8 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 1.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 2.7 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 1.5 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.1 | 2.7 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 0.8 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 2.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 2.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.7 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.1 | 11.4 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 2.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 2.3 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 1.9 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.7 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 2.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.6 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 2.7 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.5 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.1 | 2.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.5 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 9.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 0.4 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 2.9 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 1.7 | GO:0061650 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.1 | 1.6 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 3.0 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 2.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.9 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 5.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 1.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 5.5 | GO:0070405 | ammonium ion binding(GO:0070405) |
| 0.1 | 1.7 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.6 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.1 | 1.7 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 3.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.3 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 3.1 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 3.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.2 | GO:0004775 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.1 | 0.9 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.8 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 1.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 3.0 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 1.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.7 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.3 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 3.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 1.4 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 2.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 3.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 1.5 | GO:0033558 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.0 | 2.4 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 2.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 7.5 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.9 | GO:0016779 | nucleotidyltransferase activity(GO:0016779) |
| 0.0 | 0.6 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0050542 | icosanoid binding(GO:0050542) icosatetraenoic acid binding(GO:0050543) arachidonic acid binding(GO:0050544) DBD domain binding(GO:0050692) fatty acid derivative binding(GO:1901567) |
| 0.0 | 1.0 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 2.1 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 2.6 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.1 | GO:0001614 | purinergic nucleotide receptor activity(GO:0001614) extracellular ATP-gated cation channel activity(GO:0004931) nucleotide receptor activity(GO:0016502) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 1.4 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 1.8 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 18.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.5 | 6.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.3 | 34.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 17.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 3.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.2 | 6.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.2 | 7.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 8.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 10.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 8.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 8.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 10.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 12.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.2 | 14.4 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 3.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 5.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 2.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 10.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 1.2 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.1 | 9.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 2.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 4.7 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 1.7 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 3.9 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 2.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 4.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 2.8 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 2.8 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 1.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 4.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 4.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.8 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.1 | 2.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 2.0 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 1.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 2.4 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.3 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 1.7 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.4 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 9.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 2.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 5.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 3.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.3 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 22.1 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.8 | 8.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.8 | 68.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.8 | 7.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.7 | 13.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.6 | 17.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.6 | 19.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.5 | 14.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.5 | 10.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.5 | 8.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.5 | 7.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.4 | 15.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.4 | 3.3 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.4 | 6.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.3 | 9.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.3 | 6.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.3 | 11.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.3 | 7.5 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.3 | 15.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.3 | 2.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.3 | 3.9 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.3 | 4.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 3.7 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.2 | 23.0 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.2 | 1.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.2 | 3.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.2 | 3.0 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.2 | 4.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 10.1 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.2 | 4.6 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.2 | 4.0 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 9.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.2 | 3.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 3.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.2 | 4.8 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.2 | 1.5 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 3.7 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 5.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 7.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 2.0 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 9.0 | REACTOME CDT1 ASSOCIATION WITH THE CDC6 ORC ORIGIN COMPLEX | Genes involved in CDT1 association with the CDC6:ORC:origin complex |
| 0.1 | 2.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 2.1 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 3.4 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 4.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 3.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 5.3 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 2.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 2.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 2.4 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 1.0 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 4.5 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 17.2 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.1 | 1.0 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 2.0 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 2.1 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.1 | 1.8 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.5 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 11.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 2.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 0.8 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 1.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 2.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.5 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.5 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 1.7 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 1.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.6 | REACTOME HOST INTERACTIONS OF HIV FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.0 | 1.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 2.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.7 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 1.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.7 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 5.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.6 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.6 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 1.0 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 1.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 2.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 2.1 | REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | Genes involved in TRAF6 mediated induction of NFkB and MAP kinases upon TLR7/8 or 9 activation |
| 0.0 | 4.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.4 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 3.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.4 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 0.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.4 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.7 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |