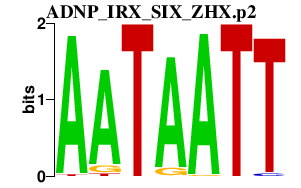
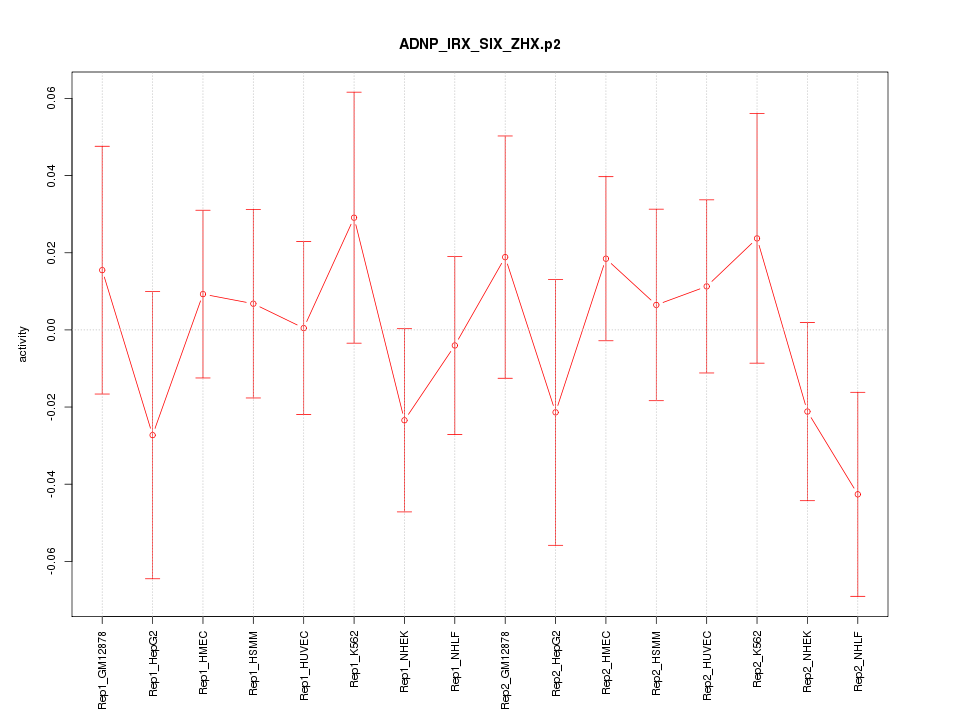
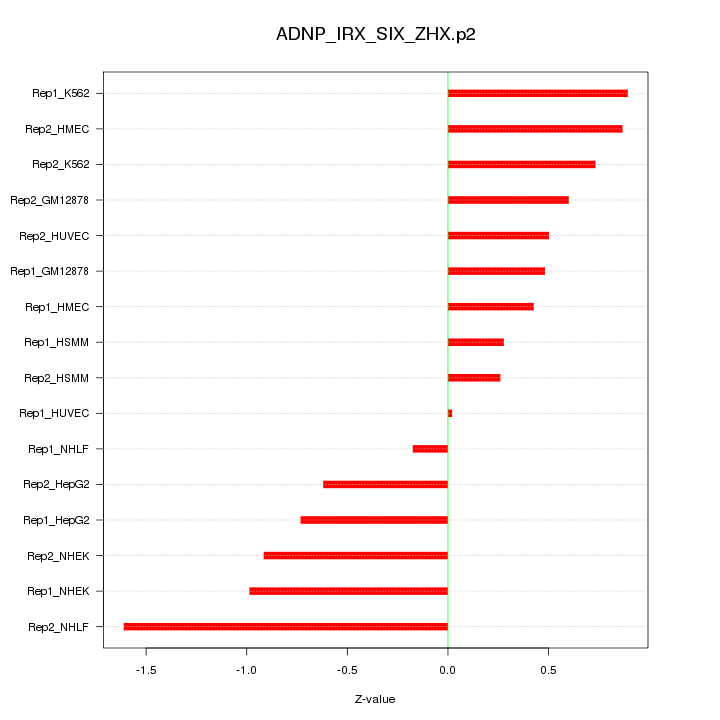
|
chrX_-_137649180
|
2.496
|
NM_033642
|
FGF13
|
fibroblast growth factor 13
|
|
chr21_-_14840506
|
2.050
|
NM_022136
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1
|
|
chr6_+_12825022
|
1.597
|
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr4_-_91979113
|
1.464
|
NM_183049
|
TMSL3
|
thymosin-like 3
|
|
chr6_+_12825818
|
1.363
|
NM_030948
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr5_-_88235624
|
1.273
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr14_-_54720101
|
1.145
|
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5
|
|
chr3_-_143230124
|
1.062
|
NM_001178141
NM_001178142
NM_006286
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr10_+_70517863
|
1.004
|
|
SRGN
|
serglycin
|
|
chr10_+_70517823
|
0.988
|
NM_002727
|
SRGN
|
serglycin
|
|
chr5_+_102493155
|
0.964
|
NM_015216
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2
|
|
chr11_-_33870411
|
0.953
|
NM_005574
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr12_+_52696908
|
0.895
|
NM_014620
NM_153693
|
HOXC4
HOXC6
|
homeobox C4
homeobox C6
|
|
chr2_+_33592445
|
0.859
|
NM_015376
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr4_-_153493133
|
0.823
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr1_+_196874759
|
0.818
|
NM_002838
NM_080921
NM_080923
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr1_-_42738535
|
0.813
|
|
TMSB4XP1
|
thymosin beta 4, X-linked pseudogene 1
|
|
chr3_+_123256902
|
0.769
|
NM_175862
|
CD86
|
CD86 molecule
|
|
chr3_-_57209319
|
0.768
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chr8_-_86477585
|
0.765
|
NM_001128829
NM_001128830
NM_001128831
NM_001738
|
CA1
|
carbonic anhydrase I
|
|
chr2_-_192420168
|
0.750
|
NM_004657
|
SDPR
|
serum deprivation response
|
|
chr12_-_69434330
|
0.749
|
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr3_-_33675484
|
0.733
|
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr1_+_196874839
|
0.724
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr14_-_105681377
|
0.643
|
|
IGHV4-31
IGHG1
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr1_+_196874867
|
0.638
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr12_-_101398503
|
0.633
|
NM_000618
NM_001111283
NM_001111285
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr20_+_29598848
|
0.627
|
|
HM13
|
histocompatibility (minor) 13
|
|
chr8_-_102872372
|
0.613
|
|
NCALD
|
neurocalcin delta
|
|
chr12_-_101398451
|
0.607
|
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr12_+_52697160
|
0.598
|
|
HOXC5
HOXC6
|
homeobox C5
homeobox C6
|
|
chr8_-_102872354
|
0.596
|
|
NCALD
|
neurocalcin delta
|
|
chr22_+_21065134
|
0.589
|
|
LOC100290481
|
immunoglobulin lambda light chain-like
|
|
chr5_+_141326620
|
0.586
|
NM_183399
|
RNF14
|
ring finger protein 14
|
|
chr12_-_101036407
|
0.582
|
NM_024057
|
NUP37
|
nucleoporin 37kDa
|
|
chr7_-_26198673
|
0.574
|
|
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1
|
|
chr12_-_15005761
|
0.572
|
NM_001175
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta
|
|
chr8_-_102872614
|
0.563
|
NM_032041
|
NCALD
|
neurocalcin delta
|
|
chrX_-_153698930
|
0.561
|
|
HMGN1P37
|
high-mobility group nucleosome binding domain 1 pseudogene 37
|
|
chr11_-_56951628
|
0.554
|
NM_017611
|
SLC43A3
|
solute carrier family 43, member 3
|
|
chr9_+_2005218
|
0.550
|
NM_003070
NM_139045
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2
|
|
chr13_+_51933228
|
0.542
|
|
CKAP2
|
cytoskeleton associated protein 2
|
|
chr3_+_174598937
|
0.541
|
NM_014932
|
NLGN1
|
neuroligin 1
|
|
chr4_+_146250978
|
0.535
|
|
ABCE1
|
ATP-binding cassette, sub-family E (OABP), member 1
|
|
chr4_+_169250281
|
0.528
|
NM_007193
|
ANXA10
|
annexin A10
|
|
chr22_+_21116283
|
0.506
|
|
|
|
|
chr3_-_168674502
|
0.505
|
NM_006217
|
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2
|
|
chr22_+_21065207
|
0.502
|
|
IGLJ3
|
immunoglobulin lambda joining 3
|
|
chr2_-_192419896
|
0.497
|
|
SDPR
|
serum deprivation response
|
|
chr6_-_33016561
|
0.486
|
NM_002118
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta
|
|
chr4_-_72868598
|
0.484
|
NM_000583
|
GC
|
group-specific component (vitamin D binding protein)
|
|
chr12_+_29267863
|
0.475
|
NM_018099
|
FAR2
|
fatty acyl CoA reductase 2
|
|
chr12_-_101398429
|
0.472
|
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr20_-_7869068
|
0.468
|
NM_017545
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1
|
|
chr11_+_113280727
|
0.460
|
NM_006028
|
HTR3B
|
5-hydroxytryptamine (serotonin) receptor 3B
|
|
chrX_+_99726445
|
0.456
|
NM_022144
|
TNMD
|
tenomodulin
|
|
chr11_+_10433241
|
0.455
|
NM_001025389
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr7_-_13992351
|
0.451
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr8_-_82557956
|
0.447
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr1_-_156923111
|
0.444
|
NM_003126
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2)
|
|
chr12_-_101398470
|
0.442
|
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr17_-_10393664
|
0.439
|
NM_001100112
NM_017534
|
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult
|
|
chr1_+_166516901
|
0.435
|
NM_005149
|
TBX19
|
T-box 19
|
|
chr1_+_35623688
|
0.428
|
|
ZMYM4
|
zinc finger, MYM-type 4
|
|
chr6_-_26343139
|
0.425
|
NM_005320
|
HIST1H1D
|
histone cluster 1, H1d
|
|
chr4_-_71751070
|
0.424
|
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides
|
|
chr17_-_10265991
|
0.418
|
NM_002472
|
MYH8
|
myosin, heavy chain 8, skeletal muscle, perinatal
|
|
chr15_-_23201740
|
0.417
|
NM_130838
|
UBE3A
|
ubiquitin protein ligase E3A
|
|
chr11_-_118405288
|
0.416
|
NM_001164280
|
SLC37A4
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4
|
|
chr6_-_33149244
|
0.416
|
NM_033554
|
HLA-DPA1
|
major histocompatibility complex, class II, DP alpha 1
|
|
chr16_+_33168191
|
0.408
|
NM_001099687
|
TP53TG3B
|
TP53 target 3B
|
|
chr12_+_49155798
|
0.405
|
|
LARP4
|
La ribonucleoprotein domain family, member 4
|
|
chr3_-_25658829
|
0.405
|
|
TOP2B
|
topoisomerase (DNA) II beta 180kDa
|
|
chr2_-_200033445
|
0.397
|
|
SATB2
|
SATB homeobox 2
|
|
chr2_-_61618165
|
0.394
|
|
XPO1
|
exportin 1 (CRM1 homolog, yeast)
|
|
chr10_-_69422391
|
0.391
|
|
HERC4
|
hect domain and RLD 4
|
|
chr16_+_7322751
|
0.388
|
NM_145891
NM_145892
NM_145893
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr7_-_36992221
|
0.387
|
NM_130442
|
ELMO1
|
engulfment and cell motility 1
|
|
chr12_-_69434650
|
0.386
|
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr12_-_101398288
|
0.386
|
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr17_-_36444626
|
0.379
|
NM_030966
|
KRTAP1-3
|
keratin associated protein 1-3
|
|
chr4_+_170817787
|
0.377
|
|
CLCN3
|
chloride channel 3
|
|
chr13_-_98757634
|
0.377
|
NM_004951
|
GPR183
|
G protein-coupled receptor 183
|
|
chr2_+_169465995
|
0.370
|
NM_001081686
NM_021176
|
G6PC2
|
glucose-6-phosphatase, catalytic, 2
|
|
chr10_+_70745615
|
0.370
|
NM_033496
|
HK1
|
hexokinase 1
|
|
chr11_-_83071084
|
0.368
|
NM_001142702
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chrX_+_28515479
|
0.366
|
NM_014271
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1
|
|
chr1_+_165565006
|
0.365
|
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr7_-_16471817
|
0.364
|
NM_015464
|
SOSTDC1
|
sclerostin domain containing 1
|
|
chr22_+_21006814
|
0.363
|
|
|
|
|
chr7_-_104696678
|
0.359
|
NM_182691
|
SRPK2
|
SRSF protein kinase 2
|
|
chr10_-_98021144
|
0.355
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr4_+_41395493
|
0.354
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr5_-_88155360
|
0.351
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chrX_-_55074125
|
0.351
|
NM_000032
NM_001037967
NM_001037968
|
ALAS2
|
aminolevulinate, delta-, synthase 2
|
|
chr1_+_165564904
|
0.350
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr7_-_132371208
|
0.349
|
|
EEF1G
|
eukaryotic translation elongation factor 1 gamma
|
|
chr2_+_33514896
|
0.346
|
NM_170672
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr7_-_25234582
|
0.345
|
NM_022150
|
NPVF
|
neuropeptide VF precursor
|
|
chr8_+_77756061
|
0.342
|
NM_024721
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr7_-_87694218
|
0.342
|
NM_198901
|
SRI
|
sorcin
|
|
chr5_+_44844724
|
0.340
|
NM_016640
|
MRPS30
|
mitochondrial ribosomal protein S30
|
|
chr1_-_157950898
|
0.338
|
NM_000567
|
CRP
|
C-reactive protein, pentraxin-related
|
|
chr2_+_237143118
|
0.337
|
NM_020311
|
CXCR7
|
chemokine (C-X-C motif) receptor 7
|
|
chr11_+_113672407
|
0.337
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr8_-_86441115
|
0.336
|
NM_001164830
|
CA1
|
carbonic anhydrase I
|
|
chr12_-_69434598
|
0.336
|
NM_130846
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr1_+_51208499
|
0.332
|
|
CDKN2C
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4)
|
|
chr14_+_66901237
|
0.330
|
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa
|
|
chr17_-_64752550
|
0.328
|
NM_080282
|
ABCA10
|
ATP-binding cassette, sub-family A (ABC1), member 10
|
|
chr12_+_14429499
|
0.327
|
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr17_-_10362583
|
0.326
|
NM_005963
|
MYH1
|
myosin, heavy chain 1, skeletal muscle, adult
|
|
chrX_-_13744933
|
0.325
|
|
GPM6B
|
glycoprotein M6B
|
|
chr13_-_45654296
|
0.325
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr7_-_26202626
|
0.324
|
|
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1
|
|
chr3_+_69895651
|
0.324
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr1_+_100957883
|
0.322
|
NM_001078
NM_080682
|
VCAM1
|
vascular cell adhesion molecule 1
|
|
chr2_-_136350404
|
0.322
|
|
MCM6
|
minichromosome maintenance complex component 6
|
|
chr18_+_31488530
|
0.320
|
NM_020474
|
GALNT1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1)
|
|
chr13_-_45654327
|
0.318
|
NM_002298
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr5_+_128468785
|
0.317
|
|
ISOC1
|
isochorismatase domain containing 1
|
|
chr6_-_22405708
|
0.316
|
NM_000948
|
PRL
|
prolactin
|
|
chr12_-_89922888
|
0.316
|
NM_004950
|
EPYC
|
epiphycan
|
|
chr11_+_113672378
|
0.314
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr3_-_166278969
|
0.314
|
NM_001041
|
SI
|
sucrase-isomaltase (alpha-glucosidase)
|
|
chr19_+_12995782
|
0.313
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr1_+_157067729
|
0.313
|
NM_002432
|
MNDA
|
myeloid cell nuclear differentiation antigen
|
|
chr17_-_37681917
|
0.312
|
NM_012448
|
STAT5B
|
signal transducer and activator of transcription 5B
|
|
chr8_+_77756077
|
0.312
|
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr5_+_157090900
|
0.312
|
NM_017872
|
THG1L
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae)
|
|
chr6_-_74284522
|
0.311
|
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1
|
|
chr1_+_198263352
|
0.311
|
NM_003822
NM_205860
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2
|
|
chr2_-_133145539
|
0.310
|
NM_001077427
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr4_-_74307637
|
0.306
|
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr12_-_8656705
|
0.306
|
NM_020661
|
AICDA
|
activation-induced cytidine deaminase
|
|
chr4_+_74825138
|
0.305
|
NM_000584
|
IL8
|
interleukin 8
|
|
chr5_-_98290117
|
0.304
|
NM_001270
|
CHD1
|
chromodomain helicase DNA binding protein 1
|
|
chrX_-_135690770
|
0.304
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chrX_-_135690699
|
0.304
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr8_+_18123456
|
0.304
|
NM_001160174
|
NAT1
|
N-acetyltransferase 1 (arylamine N-acetyltransferase)
|
|
chr2_+_172530120
|
0.303
|
|
HAT1
|
histone acetyltransferase 1
|
|
chr9_+_92629525
|
0.301
|
NM_001174167
|
SYK
|
spleen tyrosine kinase
|
|
chr2_-_170138558
|
0.301
|
NM_024622
|
FASTKD1
|
FAST kinase domains 1
|
|
chr6_-_32605898
|
0.299
|
NM_002125
|
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 5
|
|
chr11_+_63282639
|
0.299
|
|
RTN3
|
reticulon 3
|
|
chr1_+_51855343
|
0.297
|
|
OSBPL9
|
oxysterol binding protein-like 9
|
|
chrX_-_100999204
|
0.297
|
NM_032946
|
NXF5
|
nuclear RNA export factor 5
|
|
chr10_-_17211795
|
0.295
|
NM_001081
|
CUBN
|
cubilin (intrinsic factor-cobalamin receptor)
|
|
chr5_+_65408500
|
0.293
|
|
ERBB2IP
|
erbb2 interacting protein
|
|
chr2_+_102401580
|
0.290
|
NM_003853
|
IL18RAP
|
interleukin 18 receptor accessory protein
|
|
chr4_-_71751205
|
0.287
|
NM_144646
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides
|
|
chr7_-_27133163
|
0.283
|
NM_153631
|
HOXA3
|
homeobox A3
|
|
chrX_+_37524250
|
0.282
|
|
CYBB
|
cytochrome b-245, beta polypeptide
|
|
chr1_+_78858675
|
0.282
|
NM_006820
|
IFI44L
|
interferon-induced protein 44-like
|
|
chr8_-_72431274
|
0.279
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr2_-_200030944
|
0.279
|
NM_001172509
|
SATB2
|
SATB homeobox 2
|
|
chr2_-_210888010
|
0.272
|
NM_079420
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast
|
|
chr5_+_156628929
|
0.271
|
NM_014376
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr8_-_108579270
|
0.271
|
NM_001146
|
ANGPT1
|
angiopoietin 1
|
|
chr16_+_20682812
|
0.270
|
NM_005622
NM_202000
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3
|
|
chr1_+_149299530
|
0.268
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr6_+_64414389
|
0.268
|
NM_015153
|
PHF3
|
PHD finger protein 3
|
|
chr1_+_78243181
|
0.266
|
NM_007034
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4
|
|
chrX_+_100241453
|
0.266
|
NM_006733
|
CENPI
|
centromere protein I
|
|
chr1_-_51660328
|
0.264
|
NM_001159969
|
EPS15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr10_-_94247450
|
0.263
|
NM_001165946
|
IDE
|
insulin-degrading enzyme
|
|
chr1_-_111544784
|
0.261
|
NM_024901
|
DENND2D
|
DENN/MADD domain containing 2D
|
|
chr1_+_149299307
|
0.260
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr6_-_79732391
|
0.260
|
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr3_+_88270951
|
0.258
|
NM_018293
|
ZNF654
|
zinc finger protein 654
|
|
chr11_-_71424614
|
0.256
|
|
|
|
|
chr9_+_134843918
|
0.256
|
NM_001135031
NM_004188
|
GFI1B
|
growth factor independent 1B transcription repressor
|
|
chr14_+_31868229
|
0.255
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr1_+_51855133
|
0.254
|
NM_024586
NM_148906
NM_148908
NM_148909
|
OSBPL9
|
oxysterol binding protein-like 9
|
|
chr5_-_178977441
|
0.254
|
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H)
|
|
chr2_-_99412694
|
0.251
|
|
REV1
|
REV1 homolog (S. cerevisiae)
|
|
chr21_-_38955487
|
0.251
|
NM_001136154
NM_004449
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr10_+_11087259
|
0.250
|
NM_001025077
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr7_+_129057152
|
0.250
|
NM_001040110
|
NRF1
|
nuclear respiratory factor 1
|
|
chr1_-_149004852
|
0.249
|
|
CTSS
|
cathepsin S
|
|
chr13_+_32488477
|
0.248
|
NM_004795
|
KL
|
klotho
|
|
chr1_+_145892190
|
0.246
|
NM_001097616
|
GPR89C
|
G protein-coupled receptor 89C
|
|
chr1_+_115373937
|
0.245
|
NM_000549
|
TSHB
|
thyroid stimulating hormone, beta
|
|
chr3_+_120799411
|
0.245
|
NM_015900
|
PLA1A
|
phospholipase A1 member A
|
|
chr12_+_21417068
|
0.243
|
NM_000415
|
IAPP
|
islet amyloid polypeptide
|
|
chr4_-_176970407
|
0.242
|
|
GPM6A
|
glycoprotein M6A
|
|
chr11_+_113671744
|
0.241
|
NM_006169
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr3_+_179759181
|
0.238
|
NM_005832
|
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2
|
|
chr12_+_46802695
|
0.238
|
NM_001166687
NM_001166688
|
PFKM
|
phosphofructokinase, muscle
|
|
chr22_+_21559959
|
0.238
|
NM_001178126
|
IGLL5
IGLC1
|
immunoglobulin lambda-like polypeptide 5
immunoglobulin lambda constant 1 (Mcg marker)
|
|
chr1_-_239865724
|
0.235
|
NM_001821
|
CHML
|
choroideremia-like (Rab escort protein 2)
|
|
chr1_+_50347188
|
0.234
|
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr7_-_107667728
|
0.233
|
NM_001037132
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr4_-_123761615
|
0.232
|
NM_021803
|
IL21
|
interleukin 21
|
|
chr11_+_71578606
|
0.229
|
NM_000802
|
FOLR1
|
folate receptor 1 (adult)
|
|
chr15_-_48434567
|
0.228
|
|
GABPB1
|
GA binding protein transcription factor, beta subunit 1
|
|
chr2_+_161981046
|
0.226
|
|
TBR1
|
T-box, brain, 1
|
|
chr13_-_45654292
|
0.226
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr10_-_74676969
|
0.224
|
NM_015190
|
DNAJC9
|
DnaJ (Hsp40) homolog, subfamily C, member 9
|
|
chr2_-_136350472
|
0.224
|
NM_005915
|
MCM6
|
minichromosome maintenance complex component 6
|