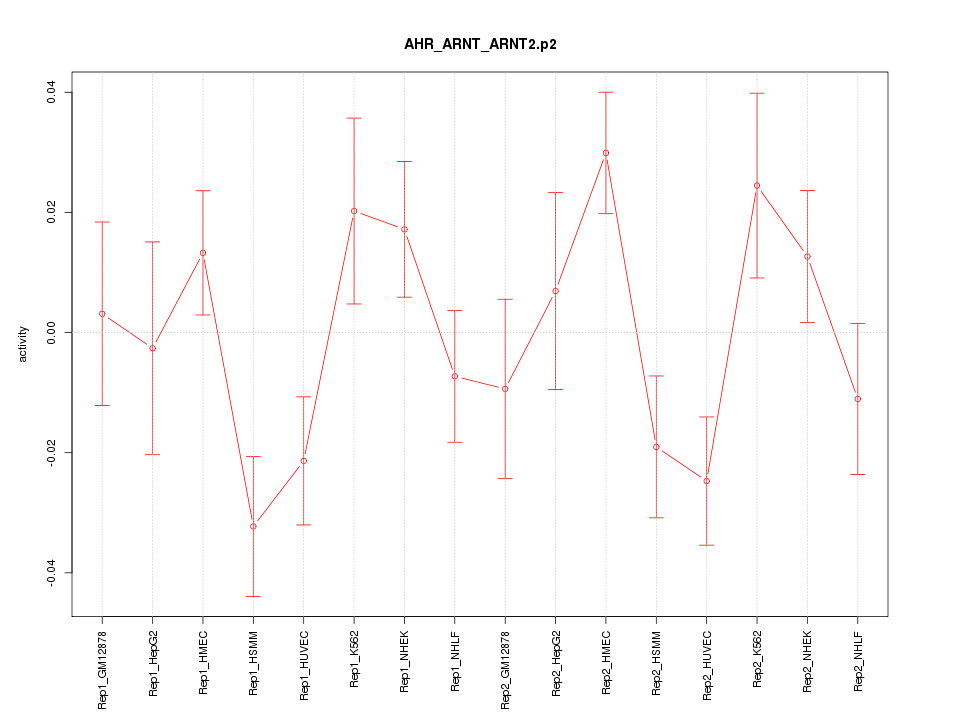
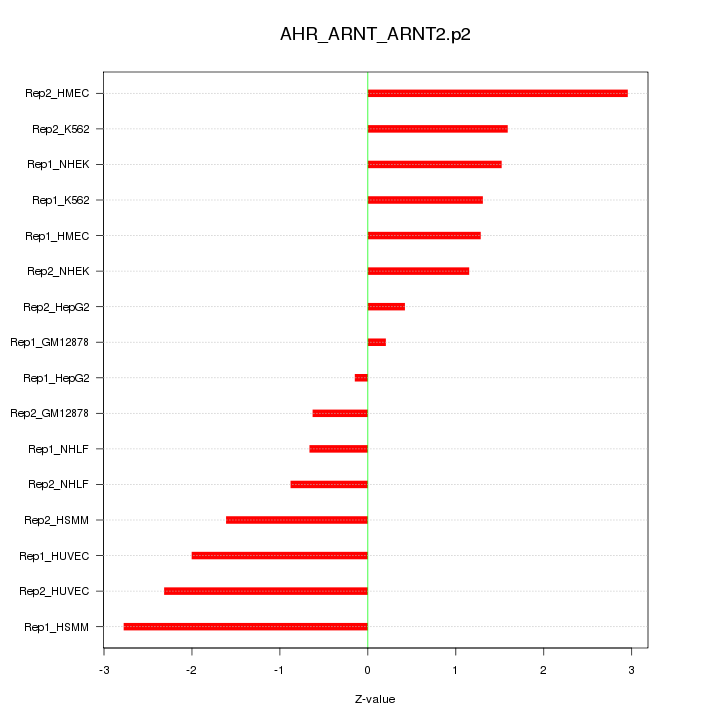
|
chr18_+_19523526
|
2.959
|
NM_001127717
NM_198129
|
LAMA3
|
laminin, alpha 3
|
|
chr2_+_203208064
|
1.990
|
NM_173511
|
FAM117B
|
family with sequence similarity 117, member B
|
|
chr12_+_4253143
|
1.477
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr6_-_10520592
|
1.452
|
NM_001032280
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr16_+_22733307
|
1.251
|
NM_006043
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr8_+_126079929
|
1.019
|
|
SQLE
|
squalene epoxidase
|
|
chr15_+_38923914
|
1.017
|
NM_001032367
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chr15_+_38923470
|
0.997
|
NM_003710
NM_181642
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chr1_-_6585268
|
0.988
|
|
KLHL21
|
kelch-like 21 (Drosophila)
|
|
chr10_-_103869923
|
0.976
|
NM_001113407
|
LDB1
|
LIM domain binding 1
|
|
chr1_-_43196911
|
0.965
|
|
SLC2A1
|
solute carrier family 2 (facilitated glucose transporter), member 1
|
|
chr1_-_58815415
|
0.942
|
|
TACSTD2
|
tumor-associated calcium signal transducer 2
|
|
chr15_-_35178888
|
0.939
|
NM_172316
|
MEIS2
|
Meis homeobox 2
|
|
chr15_+_81906983
|
0.896
|
NM_003027
|
SH3GL3
|
SH3-domain GRB2-like 3
|
|
chr7_-_91601613
|
0.852
|
|
CYP51A1
|
cytochrome P450, family 51, subfamily A, polypeptide 1
|
|
chrX_+_37429927
|
0.844
|
NM_021083
|
XK
|
X-linked Kx blood group (McLeod syndrome)
|
|
chr15_-_73719546
|
0.833
|
NM_018285
|
IMP3
|
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast)
|
|
chr8_+_126079900
|
0.832
|
NM_003129
|
SQLE
|
squalene epoxidase
|
|
chr15_-_66900266
|
0.822
|
|
ANP32A
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A
|
|
chr15_+_50098730
|
0.816
|
|
MAPK6
|
mitogen-activated protein kinase 6
|
|
chr3_+_195336625
|
0.814
|
NM_005524
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chrX_+_109132994
|
0.803
|
NM_032227
|
TMEM164
|
transmembrane protein 164
|
|
chr3_+_195336631
|
0.802
|
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr15_+_50098724
|
0.780
|
|
MAPK6
|
mitogen-activated protein kinase 6
|
|
chr20_-_33335919
|
0.772
|
NM_181468
|
EIF6
|
eukaryotic translation initiation factor 6
|
|
chr15_-_61461127
|
0.772
|
NM_001218
NM_206925
|
CA12
|
carbonic anhydrase XII
|
|
chr7_-_91601711
|
0.769
|
|
CYP51A1
|
cytochrome P450, family 51, subfamily A, polypeptide 1
|
|
chr1_+_1396975
|
0.762
|
|
ATAD3B
|
ATPase family, AAA domain containing 3B
|
|
chr19_-_50601406
|
0.754
|
NM_001142502
|
PPP1R13L
|
protein phosphatase 1, regulatory (inhibitor) subunit 13 like
|
|
chr1_+_1397021
|
0.728
|
NM_031921
|
ATAD3B
|
ATPase family, AAA domain containing 3B
|
|
chr8_+_126511739
|
0.723
|
NM_025195
|
TRIB1
|
tribbles homolog 1 (Drosophila)
|
|
chr20_+_20296744
|
0.722
|
NM_002196
|
INSM1
|
insulinoma-associated 1
|
|
chr19_-_614158
|
0.702
|
|
RNF126
|
ring finger protein 126
|
|
chr7_-_91601764
|
0.696
|
NM_000786
|
CYP51A1
|
cytochrome P450, family 51, subfamily A, polypeptide 1
|
|
chr15_-_61460967
|
0.682
|
|
CA12
|
carbonic anhydrase XII
|
|
chr20_-_33335703
|
0.678
|
|
EIF6
|
eukaryotic translation initiation factor 6
|
|
chr6_+_15353681
|
0.658
|
|
JARID2
|
jumonji, AT rich interactive domain 2
|
|
chr19_-_614155
|
0.655
|
|
RNF126
|
ring finger protein 126
|
|
chr12_-_44948800
|
0.648
|
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chrX_+_152608159
|
0.635
|
NM_001142806
|
SLC6A8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8
|
|
chr19_-_614219
|
0.621
|
NM_194460
|
RNF126
|
ring finger protein 126
|
|
chr8_+_31616809
|
0.610
|
NM_013962
|
NRG1
|
neuregulin 1
|
|
chr1_-_6585485
|
0.606
|
NM_014851
|
KLHL21
|
kelch-like 21 (Drosophila)
|
|
chrX_+_109132507
|
0.603
|
NM_017698
|
TMEM164
|
transmembrane protein 164
|
|
chr6_+_43592812
|
0.601
|
|
POLR1C
|
polymerase (RNA) I polypeptide C, 30kDa
|
|
chr6_+_125517033
|
0.591
|
NM_001003395
|
TPD52L1
|
tumor protein D52-like 1
|
|
chr16_+_22732982
|
0.589
|
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr6_+_34312667
|
0.587
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr15_+_50098700
|
0.576
|
NM_002748
|
MAPK6
|
mitogen-activated protein kinase 6
|
|
chr3_-_191321367
|
0.574
|
|
LEPREL1
|
leprecan-like 1
|
|
chr7_+_138695537
|
0.572
|
|
LUC7L2
|
LUC7-like 2 (S. cerevisiae)
|
|
chr1_+_199519202
|
0.571
|
NM_000299
NM_001005337
|
PKP1
|
plakophilin 1 (ectodermal dysplasia/skin fragility syndrome)
|
|
chr19_-_614195
|
0.571
|
|
RNF126
|
ring finger protein 126
|
|
chr22_+_20101661
|
0.565
|
NM_015094
|
HIC2
|
hypermethylated in cancer 2
|
|
chr19_-_51983024
|
0.556
|
|
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5
|
|
chr6_+_43592758
|
0.553
|
NM_004875
NM_203290
|
POLR1C
|
polymerase (RNA) I polypeptide C, 30kDa
|
|
chr15_+_99237527
|
0.548
|
NM_000693
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3
|
|
chr7_-_105712646
|
0.546
|
NM_005746
|
NAMPT
|
nicotinamide phosphoribosyltransferase
|
|
chr20_+_43468617
|
0.531
|
NM_001048223
NM_001048224
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chrX_+_48265107
|
0.518
|
NM_006579
|
EBP
|
emopamil binding protein (sterol isomerase)
|
|
chr15_+_99237555
|
0.504
|
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3
|
|
chr15_-_35179185
|
0.498
|
|
MEIS2
|
Meis homeobox 2
|
|
chr6_+_43846735
|
0.495
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr9_-_98841186
|
0.491
|
NM_001333
|
CTSL2
|
cathepsin L2
|
|
chr6_+_34312625
|
0.491
|
NM_145901
NM_145902
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr10_-_104168890
|
0.489
|
NM_002779
|
PSD
|
pleckstrin and Sec7 domain containing
|
|
chr22_+_18485068
|
0.485
|
|
RANBP1
|
RAN binding protein 1
|
|
chr6_+_125516499
|
0.482
|
NM_001003396
NM_001003397
NM_003287
|
TPD52L1
|
tumor protein D52-like 1
|
|
chr17_+_77289804
|
0.481
|
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10
|
|
chr11_-_87548199
|
0.481
|
NM_022337
|
RAB38
|
RAB38, member RAS oncogene family
|
|
chr1_-_232681471
|
0.480
|
NM_005646
|
TARBP1
|
TAR (HIV-1) RNA binding protein 1
|
|
chrX_-_48661226
|
0.474
|
NM_006875
|
PIM2
|
pim-2 oncogene
|
|
chr9_-_102154915
|
0.468
|
NM_017746
|
TEX10
|
testis expressed 10
|
|
chr10_-_104168559
|
0.463
|
|
PSD
|
pleckstrin and Sec7 domain containing
|
|
chr7_+_99865340
|
0.456
|
|
MEPCE
|
methylphosphate capping enzyme
|
|
chrX_-_18282698
|
0.452
|
NM_006089
|
SCML2
|
sex comb on midleg-like 2 (Drosophila)
|
|
chr7_-_91601972
|
0.448
|
NM_001146152
|
CYP51A1
|
cytochrome P450, family 51, subfamily A, polypeptide 1
|
|
chr6_+_30402203
|
0.445
|
|
|
|
|
chr21_+_34367678
|
0.445
|
NM_032476
NM_006933
|
MRPS6
SLC5A3
|
mitochondrial ribosomal protein S6
solute carrier family 5 (sodium/myo-inositol cotransporter), member 3
|
|
chr4_-_83569476
|
0.445
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chrX_-_137621060
|
0.444
|
|
FGF13
|
fibroblast growth factor 13
|
|
chrX_-_138114846
|
0.439
|
NM_001139498
NM_001139500
NM_001139501
|
FGF13
|
fibroblast growth factor 13
|
|
chr1_+_1437400
|
0.437
|
|
ATAD3A
ATAD3B
|
ATPase family, AAA domain containing 3A
ATPase family, AAA domain containing 3B
|
|
chr11_-_114880275
|
0.437
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chr1_-_54076521
|
0.435
|
|
TMEM48
|
transmembrane protein 48
|
|
chr5_-_134397470
|
0.433
|
|
PITX1
|
paired-like homeodomain 1
|
|
chr17_+_52517529
|
0.431
|
NM_003488
|
AKAP1
|
A kinase (PRKA) anchor protein 1
|
|
chr6_-_86409415
|
0.430
|
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr10_+_21863099
|
0.429
|
NM_001195627
NM_001195628
NM_001195630
NM_004641
|
MLLT10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10
|
|
chr16_+_31098962
|
0.429
|
|
FUS
|
fused in sarcoma
|
|
chr17_+_15789147
|
0.429
|
|
ADORA2B
|
adenosine A2b receptor
|
|
chr6_-_10523424
|
0.426
|
NM_003220
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr9_+_4975082
|
0.420
|
|
JAK2
|
Janus kinase 2
|
|
chrX_-_109447970
|
0.418
|
NM_001025580
NM_015365
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr7_+_138695241
|
0.416
|
|
LUC7L2
|
LUC7-like 2 (S. cerevisiae)
|
|
chrX_-_137621492
|
0.414
|
NM_004114
|
FGF13
|
fibroblast growth factor 13
|
|
chr6_+_135544170
|
0.413
|
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr3_+_25806552
|
0.413
|
NM_001145391
NM_017897
|
OXSM
|
3-oxoacyl-ACP synthase, mitochondrial
|
|
chr17_+_34114924
|
0.412
|
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6
|
|
chr1_-_54076695
|
0.411
|
NM_001168551
NM_018087
|
TMEM48
|
transmembrane protein 48
|
|
chr1_-_38046433
|
0.410
|
|
YRDC
|
yrdC domain containing (E. coli)
|
|
chr22_+_19601721
|
0.409
|
|
CRKL
|
v-crk sarcoma virus CT10 oncogene homolog (avian)-like
|
|
chr6_+_34312641
|
0.409
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr17_+_77582774
|
0.407
|
NM_005052
|
RAC3
|
ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3)
|
|
chrX_+_153644300
|
0.407
|
|
DKC1
|
dyskeratosis congenita 1, dyskerin
|
|
chr1_+_1437734
|
0.403
|
NM_001170536
|
ATAD3A
|
ATPase family, AAA domain containing 3A
|
|
chr6_+_135544214
|
0.403
|
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr6_+_135544138
|
0.401
|
NM_001130172
NM_001130173
NM_001161656
NM_001161657
NM_001161658
NM_001161659
NM_001161660
NM_005375
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr17_-_37328755
|
0.400
|
|
ACLY
|
ATP citrate lyase
|
|
chr12_-_31370305
|
0.400
|
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr20_-_33335985
|
0.400
|
NM_002212
NM_181466
|
EIF6
|
eukaryotic translation initiation factor 6
|
|
chr5_+_154115046
|
0.397
|
|
LARP1
|
La ribonucleoprotein domain family, member 1
|
|
chr17_+_69939806
|
0.395
|
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C
|
|
chr12_-_74710741
|
0.394
|
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr1_-_38046439
|
0.394
|
NM_024640
|
YRDC
|
yrdC domain containing (E. coli)
|
|
chrX_-_109447954
|
0.393
|
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr15_-_66900143
|
0.393
|
|
ANP32A
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A
|
|
chr22_+_18485057
|
0.390
|
|
RANBP1
|
RAN binding protein 1
|
|
chr20_+_43468046
|
0.390
|
NM_001197139
NM_001048221
NM_001048222
NM_001197140
NM_018478
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr7_+_138695236
|
0.390
|
|
LUC7L2
|
LUC7-like 2 (S. cerevisiae)
|
|
chr17_-_37328679
|
0.388
|
|
ACLY
|
ATP citrate lyase
|
|
chr2_+_219283751
|
0.387
|
NM_014640
|
TTLL4
|
tubulin tyrosine ligase-like family, member 4
|
|
chr16_+_31098963
|
0.386
|
|
FUS
|
fused in sarcoma
|
|
chr17_+_69939130
|
0.386
|
NM_022036
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C
|
|
chr6_+_34312633
|
0.385
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr19_+_1358750
|
0.383
|
|
DAZAP1
|
DAZ associated protein 1
|
|
chr7_-_95789204
|
0.383
|
NM_001160210
NM_014251
|
SLC25A13
|
solute carrier family 25, member 13 (citrin)
|
|
chr19_+_43447262
|
0.382
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr14_+_100262916
|
0.378
|
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chr2_+_28971007
|
0.378
|
NM_015131
|
WDR43
|
WD repeat domain 43
|
|
chr1_-_165789640
|
0.377
|
NM_003851
|
CREG1
|
cellular repressor of E1A-stimulated genes 1
|
|
chr5_-_176763991
|
0.377
|
|
F12
|
coagulation factor XII (Hageman factor)
|
|
chr7_+_138695209
|
0.377
|
|
LUC7L2
|
LUC7-like 2 (S. cerevisiae)
|
|
chr4_+_4439705
|
0.375
|
|
D4S234E
|
DNA segment on chromosome 4 (unique) 234 expressed sequence
|
|
chr6_-_86409283
|
0.375
|
NM_001159674
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr4_-_185984045
|
0.374
|
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr1_+_35019376
|
0.373
|
NM_024009
|
GJB3
|
gap junction protein, beta 3, 31kDa
|
|
chrX_+_130984912
|
0.372
|
NM_001042453
NM_016542
|
MST4
|
serine/threonine protein kinase MST4
|
|
chr17_-_77649393
|
0.371
|
NM_004104
|
FASN
|
fatty acid synthase
|
|
chr5_+_6686499
|
0.369
|
NM_001047
|
SRD5A1
|
steroid-5-alpha-reductase, alpha polypeptide 1 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 1)
|
|
chr19_+_50663089
|
0.369
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr12_-_31370351
|
0.369
|
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr1_-_165789586
|
0.369
|
|
CREG1
|
cellular repressor of E1A-stimulated genes 1
|
|
chr1_-_53566171
|
0.367
|
NM_001018054
NM_004631
NM_017522
|
LRP8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor
|
|
chr12_-_31370387
|
0.367
|
NM_001135811
NM_021238
NM_001135812
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr19_+_50601306
|
0.367
|
NM_012099
|
CD3EAP
|
CD3e molecule, epsilon associated protein
|
|
chr5_-_134942867
|
0.366
|
NM_004887
|
CXCL14
|
chemokine (C-X-C motif) ligand 14
|
|
chr17_+_15788955
|
0.366
|
NM_000676
|
ADORA2B
|
adenosine A2b receptor
|
|
chr22_+_19601723
|
0.362
|
|
CRKL
|
v-crk sarcoma virus CT10 oncogene homolog (avian)-like
|
|
chr1_-_38046270
|
0.360
|
|
YRDC
|
yrdC domain containing (E. coli)
|
|
chr5_+_6686531
|
0.360
|
|
SRD5A1
|
steroid-5-alpha-reductase, alpha polypeptide 1 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 1)
|
|
chr1_-_165789630
|
0.359
|
|
CREG1
|
cellular repressor of E1A-stimulated genes 1
|
|
chr10_-_105442868
|
0.357
|
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr4_-_56996400
|
0.356
|
NM_002703
|
PPAT
|
phosphoribosyl pyrophosphate amidotransferase
|
|
chr17_-_37328716
|
0.355
|
|
ACLY
|
ATP citrate lyase
|
|
chr1_+_54184595
|
0.352
|
NM_052940
|
LRRC42
|
leucine rich repeat containing 42
|
|
chr1_-_38046410
|
0.352
|
|
YRDC
|
yrdC domain containing (E. coli)
|
|
chr17_-_37328764
|
0.351
|
NM_001096
NM_198830
|
ACLY
|
ATP citrate lyase
|
|
chr10_+_21863272
|
0.350
|
|
MLLT10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10
|
|
chr16_+_15397120
|
0.350
|
|
MPV17L
|
MPV17 mitochondrial membrane protein-like
|
|
chr4_-_39655554
|
0.347
|
|
PDS5A
|
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae)
|
|
chr7_+_16759845
|
0.347
|
NM_014399
|
TSPAN13
|
tetraspanin 13
|
|
chr2_+_169021039
|
0.347
|
NM_203463
|
LASS6
|
LAG1 homolog, ceramide synthase 6
|
|
chr20_-_10602139
|
0.346
|
|
JAG1
|
jagged 1
|
|
chr8_+_102574012
|
0.344
|
|
GRHL2
|
grainyhead-like 2 (Drosophila)
|
|
chr7_+_99865189
|
0.343
|
NM_019606
|
MEPCE
|
methylphosphate capping enzyme
|
|
chr7_-_105712565
|
0.343
|
|
NAMPT
|
nicotinamide phosphoribosyltransferase
|
|
chr7_+_154720547
|
0.343
|
|
INSIG1
|
insulin induced gene 1
|
|
chr17_-_44058612
|
0.342
|
NM_024017
|
HOXB9
|
homeobox B9
|
|
chr6_-_119712596
|
0.341
|
NM_005907
|
MAN1A1
|
mannosidase, alpha, class 1A, member 1
|
|
chr7_+_138695144
|
0.341
|
NM_016019
|
LUC7L2
|
LUC7-like 2 (S. cerevisiae)
|
|
chr12_-_119116848
|
0.339
|
NM_006836
|
GCN1L1
|
GCN1 general control of amino-acid synthesis 1-like 1 (yeast)
|
|
chrX_+_19271931
|
0.337
|
NM_000284
NM_001173454
NM_001173455
NM_001173456
|
PDHA1
|
pyruvate dehydrogenase (lipoamide) alpha 1
|
|
chr7_+_2360954
|
0.336
|
NM_001037283
NM_003751
|
EIF3B
|
eukaryotic translation initiation factor 3, subunit B
|
|
chr5_+_49998528
|
0.336
|
NM_001178056
NM_024615
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr1_+_1437358
|
0.334
|
NM_001170535
NM_018188
|
ATAD3A
|
ATPase family, AAA domain containing 3A
|
|
chr17_+_77289704
|
0.331
|
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10
|
|
chrX_+_153644210
|
0.331
|
NM_001142463
NM_001363
|
DKC1
|
dyskeratosis congenita 1, dyskerin
|
|
chr4_-_76817582
|
0.330
|
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2
|
|
chr8_+_110621092
|
0.330
|
NM_198120
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9
|
|
chr7_+_115637795
|
0.329
|
NM_015641
|
TES
|
testis derived transcript (3 LIM domains)
|
|
chr7_+_2361051
|
0.327
|
|
EIF3B
|
eukaryotic translation initiation factor 3, subunit B
|
|
chr11_-_78829342
|
0.327
|
NM_001098816
|
ODZ4
|
odz, odd Oz/ten-m homolog 4 (Drosophila)
|
|
chr4_-_76817587
|
0.325
|
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2
|
|
chr7_+_2361024
|
0.324
|
|
EIF3B
|
eukaryotic translation initiation factor 3, subunit B
|
|
chr20_-_10602429
|
0.324
|
NM_000214
|
JAG1
|
jagged 1
|
|
chr9_-_99994704
|
0.324
|
NM_052820
|
CORO2A
|
coronin, actin binding protein, 2A
|
|
chr16_+_23754822
|
0.324
|
|
PRKCB
|
protein kinase C, beta
|
|
chr16_+_15397087
|
0.323
|
NM_001128423
NM_173803
|
MPV17L
|
MPV17 mitochondrial membrane protein-like
|
|
chr4_+_38341982
|
0.323
|
NM_016531
|
KLF3
|
Kruppel-like factor 3 (basic)
|
|
chr2_+_181553783
|
0.322
|
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 (UBC4/5 homolog, yeast)
|
|
chr8_+_110621485
|
0.321
|
NM_004215
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9
|
|
chr6_-_86409354
|
0.321
|
NM_001159673
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr17_-_38081604
|
0.321
|
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3
|
|
chr1_+_219119623
|
0.319
|
|
HLX
|
H2.0-like homeobox
|
|
chr22_-_19685998
|
0.318
|
|
THAP7
|
THAP domain containing 7
|
|
chr16_+_31098971
|
0.318
|
|
FUS
|
fused in sarcoma
|
|
chr1_+_65386559
|
0.318
|
|
AK4
|
adenylate kinase 4
|
|
chr3_-_52065130
|
0.317
|
NM_001947
|
DUSP7
|
dual specificity phosphatase 7
|
|
chr22_+_18485076
|
0.317
|
|
RANBP1
|
RAN binding protein 1
|