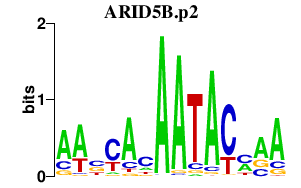
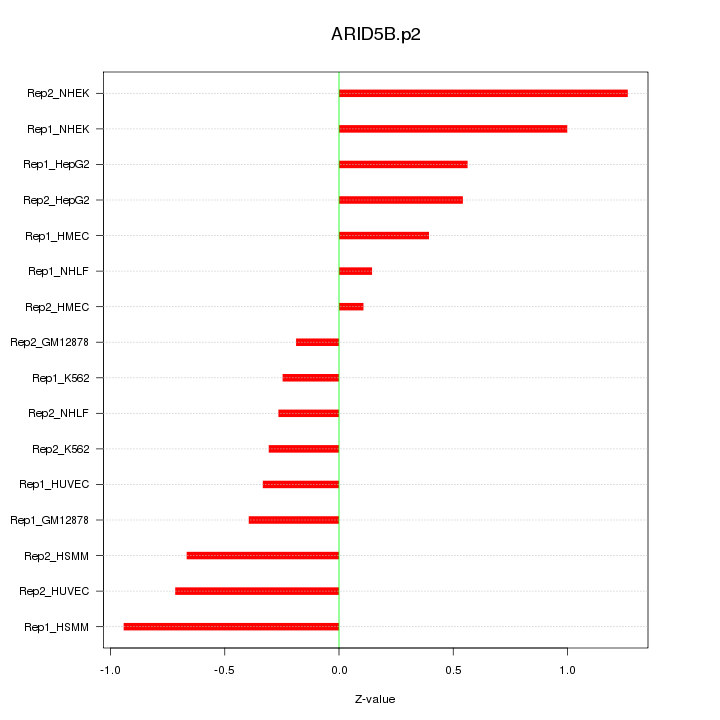
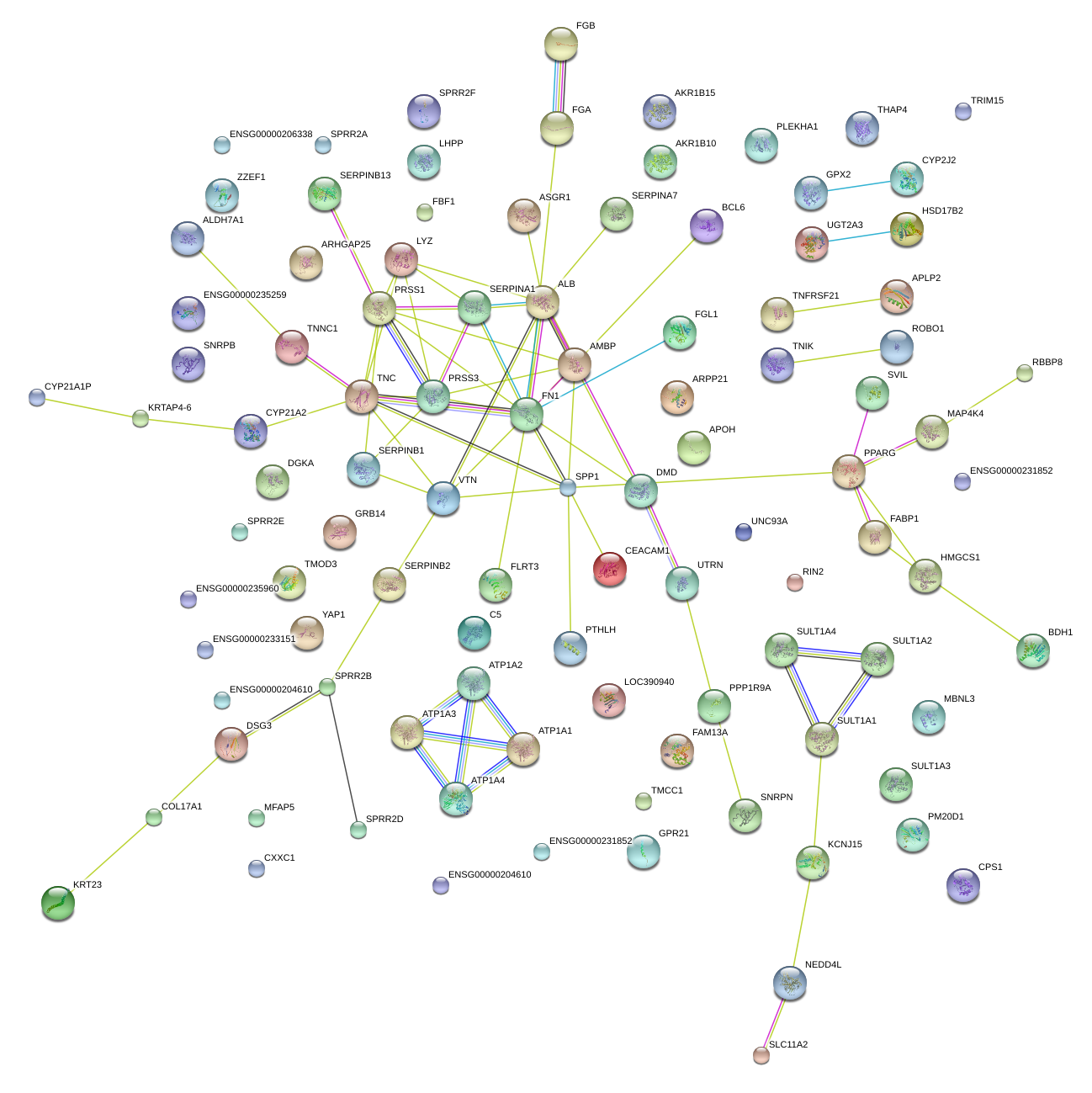
|
chr3_-_79899748
|
1.076
|
NM_002941
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr10_+_102113410
|
1.010
|
|
|
|
|
chr14_-_64479198
|
0.913
|
NM_002083
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal)
|
|
chrX_-_131375217
|
0.889
|
NM_001170701
NM_001170702
NM_001170703
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr17_-_61655915
|
0.776
|
NM_000042
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I)
|
|
chr2_-_215949566
|
0.760
|
|
FN1
|
fibronectin 1
|
|
chr11_+_101488380
|
0.740
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr20_-_14266200
|
0.621
|
NM_013281
NM_198391
|
FLRT3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr3_-_131081910
|
0.582
|
NM_001017395
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr9_-_122852356
|
0.539
|
NM_001735
|
C5
|
complement component 5
|
|
chr3_-_188946168
|
0.524
|
NM_001706
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr15_+_49988882
|
0.497
|
|
TMOD3
|
tropomodulin 3 (ubiquitous)
|
|
chr4_+_155707089
|
0.465
|
|
FGB
|
fibrinogen beta chain
|
|
chr1_-_151280201
|
0.459
|
NM_006945
|
SPRR2D
|
small proline-rich protein 2D
|
|
chr1_-_151352587
|
0.458
|
NM_001014450
|
SPRR2A
SPRR2F
|
small proline-rich protein 2A
small proline-rich protein 2F
|
|
chr6_+_30238961
|
0.453
|
NM_033229
|
TRIM15
|
tripartite motif containing 15
|
|
chr14_-_93914634
|
0.447
|
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr1_-_151310707
|
0.438
|
NM_001017418
|
SPRR2B
|
small proline-rich protein 2B
|
|
chrM_+_12283
|
0.421
|
|
|
|
|
chr5_-_125956996
|
0.419
|
|
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1
|
|
chr6_+_167624792
|
0.405
|
NM_001143947
NM_018974
|
UNC93A
|
unc-93 homolog A (C. elegans)
|
|
chr4_+_89115862
|
0.397
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr12_-_28016182
|
0.393
|
NM_002820
NM_198965
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr16_+_80626456
|
0.393
|
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chrX_-_105169370
|
0.378
|
NM_000354
|
SERPINA7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7
|
|
chr18_+_59705918
|
0.377
|
NM_001143818
NM_002575
|
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2
|
|
chr18_+_53865455
|
0.372
|
NM_001144964
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr10_-_29963906
|
0.370
|
NM_021738
|
SVIL
|
supervillin
|
|
chr4_+_89115825
|
0.346
|
NM_000582
NM_001040058
NM_001040060
|
SPP1
|
secreted phosphoprotein 1
|
|
chr4_+_74488910
|
0.326
|
|
ALB
|
albumin
|
|
chr16_+_80626459
|
0.322
|
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chr2_+_68815416
|
0.319
|
NM_001007231
NM_001166277
|
ARHGAP25
|
Rho GTPase activating protein 25
|
|
chr2_-_88208650
|
0.310
|
NM_001443
|
FABP1
|
fatty acid binding protein 1, liver
|
|
chr10_+_124135574
|
0.308
|
NM_001195608
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr7_+_133862883
|
0.303
|
NM_020299
|
AKR1B10
|
aldo-keto reductase family 1, member B10 (aldose reductase)
|
|
chr16_+_80626402
|
0.301
|
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chr16_+_29378694
|
0.297
|
NM_001017390
NM_177552
|
SULT1A4
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3
|
|
chr4_-_69852090
|
0.294
|
NM_024743
|
UGT2A3
|
UDP glucuronosyltransferase 2 family, polypeptide A3
|
|
chr15_+_22619886
|
0.286
|
NM_022806
NM_022807
NM_022808
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N
|
|
chr12_+_54611212
|
0.285
|
NM_001345
|
DGKA
|
diacylglycerol kinase, alpha 80kDa
|
|
chr17_-_23721351
|
0.282
|
|
VTN
|
vitronectin
|
|
chr17_-_23721420
|
0.279
|
|
VTN
|
vitronectin
|
|
chr16_+_30118036
|
0.278
|
NM_001017390
NM_177552
|
SULT1A4
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3
|
|
chr8_-_17787342
|
0.274
|
|
FGL1
|
fibrinogen-like 1
|
|
chr10_-_105835525
|
0.272
|
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr9_+_124836666
|
0.263
|
NM_005294
|
GPR21
|
G protein-coupled receptor 21
|
|
chr6_-_47361921
|
0.262
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr11_+_129484540
|
0.257
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr3_-_198784590
|
0.248
|
NM_203315
|
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1
|
|
chr2_+_101874765
|
0.241
|
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr6_-_47362102
|
0.239
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr1_-_60164912
|
0.233
|
NM_000775
|
CYP2J2
|
cytochrome P450, family 2, subfamily J, polypeptide 2
|
|
chr7_+_94377145
|
0.228
|
NM_001166161
NM_001166162
NM_001166163
|
PPP1R9A
|
protein phosphatase 1, regulatory (inhibitor) subunit 9A
|
|
chr1_-_151333624
|
0.222
|
NM_001024209
|
SPRR2E
|
small proline-rich protein 2E
|
|
chr2_-_242205556
|
0.215
|
NM_001164356
|
THAP4
|
THAP domain containing 4
|
|
chr17_-_36347197
|
0.211
|
NM_015515
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr6_-_2785682
|
0.211
|
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1
|
|
chr19_-_47723183
|
0.207
|
|
CEACAM1
|
carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein)
|
|
chr1_+_116727514
|
0.203
|
NM_001160234
|
ATP1A1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide
|
|
chr18_+_18767275
|
0.201
|
NM_002894
|
RBBP8
|
retinoblastoma binding protein 8
|
|
chr12_-_49689295
|
0.199
|
NM_001174130
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2
|
|
chr10_-_105835609
|
0.197
|
NM_000494
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr5_-_43326922
|
0.197
|
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble)
|
|
chr4_+_74500748
|
0.192
|
|
ALB
|
albumin
|
|
chrX_-_33267646
|
0.190
|
NM_000109
|
DMD
|
dystrophin
|
|
chr2_-_165133239
|
0.188
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr18_+_59405513
|
0.188
|
NM_012397
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13
|
|
chr9_-_116920232
|
0.182
|
NM_002160
|
TNC
|
tenascin C
|
|
chr9_+_33740463
|
0.179
|
NM_001197097
NM_007343
NM_001197098
|
PRSS3
|
protease, serine, 3
|
|
chr17_-_7023378
|
0.179
|
NM_001197216
NM_001671
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chr4_-_155731288
|
0.173
|
NM_000508
NM_021871
|
FGA
|
fibrinogen alpha chain
|
|
chr18_-_28247186
|
0.172
|
|
|
|
|
chr4_-_90197140
|
0.170
|
NM_014883
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr4_+_74491272
|
0.167
|
|
ALB
|
albumin
|
|
chr2_+_211166323
|
0.164
|
NM_001122634
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial
|
|
chr9_-_115877104
|
0.161
|
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr18_+_27281729
|
0.160
|
NM_001944
|
DSG3
|
desmoglein 3
|
|
chr12_+_68028397
|
0.160
|
NM_000239
|
LYZ
|
lysozyme
|
|
chr19_+_48772791
|
0.159
|
NM_001193621
|
LOC390940
|
hypothetical protein LOC390940
|
|
chr12_-_8706533
|
0.156
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr3_+_12367993
|
0.156
|
NM_015869
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr4_+_89115892
|
0.152
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr10_+_126140330
|
0.152
|
NM_001167880
NM_022126
|
LHPP
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase
|
|
chr21_+_38550533
|
0.151
|
NM_002243
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15
|
|
chr20_+_19863716
|
0.149
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr19_+_61151009
|
0.149
|
NM_176811
|
NLRP8
|
NLR family, pyrin domain containing 8
|
|
chr11_+_43622348
|
0.147
|
|
|
|
|
chr1_-_245120900
|
0.146
|
|
AHCTF1
|
AT hook containing transcription factor 1
|
|
chr5_+_176449156
|
0.144
|
NM_022963
|
FGFR4
|
fibroblast growth factor receptor 4
|
|
chr1_-_195818927
|
0.143
|
|
DENND1B
|
DENN/MADD domain containing 1B
|
|
chr4_+_75449720
|
0.134
|
NM_001432
|
EREG
|
epiregulin
|
|
chr22_+_38903825
|
0.134
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr6_-_32247134
|
0.130
|
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha)
|
|
chr4_+_187424111
|
0.129
|
NM_000128
|
F11
|
coagulation factor XI
|
|
chr7_-_87180499
|
0.126
|
NM_000927
|
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1
|
|
chr1_+_13784553
|
0.126
|
NM_001006624
NM_001006625
|
PDPN
|
podoplanin
|
|
chr18_-_59480082
|
0.125
|
NM_006919
|
SERPINB3
SERPINB4
|
serpin peptidase inhibitor, clade B (ovalbumin), member 3
serpin peptidase inhibitor, clade B (ovalbumin), member 4
|
|
chr2_+_128644510
|
0.124
|
|
UGGT1
|
UDP-glucose glycoprotein glucosyltransferase 1
|
|
chr22_+_19458378
|
0.121
|
NM_000185
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1
|
|
chr2_+_101874809
|
0.119
|
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr17_-_7023283
|
0.118
|
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chr1_+_20837001
|
0.118
|
|
|
|
|
chr18_-_59462469
|
0.118
|
NM_002974
|
SERPINB4
|
serpin peptidase inhibitor, clade B (ovalbumin), member 4
|
|
chr19_-_56223095
|
0.117
|
NM_001167605
NM_006853
|
KLK11
|
kallikrein-related peptidase 11
|
|
chr17_+_54587882
|
0.117
|
|
PRR11
|
proline rich 11
|
|
chr6_-_64087806
|
0.116
|
NM_001143940
NM_016571
|
LGSN
|
lengsin, lens protein with glutamine synthetase domain
|
|
chr11_+_57123927
|
0.116
|
|
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1
|
|
chr17_-_53847937
|
0.115
|
|
RNF43
|
ring finger protein 43
|
|
chr2_+_113591940
|
0.115
|
NM_000577
NM_173841
NM_173843
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr1_-_58920315
|
0.115
|
|
|
|
|
chr7_+_75892187
|
0.113
|
NM_001110354
|
ZP3
|
zona pellucida glycoprotein 3 (sperm receptor)
|
|
chr17_+_58440629
|
0.111
|
NM_025185
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2
|
|
chr14_-_30927931
|
0.111
|
NM_015473
|
HEATR5A
|
HEAT repeat containing 5A
|
|
chr6_+_64344307
|
0.111
|
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr6_+_131508153
|
0.109
|
NM_016377
|
AKAP7
|
A kinase (PRKA) anchor protein 7
|
|
chr8_+_104900591
|
0.107
|
NM_014677
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr1_+_92405196
|
0.106
|
NM_015237
|
KIAA1107
|
KIAA1107
|
|
chr18_-_44723116
|
0.106
|
NM_001190823
|
SMAD7
|
SMAD family member 7
|
|
chr10_-_118670968
|
0.103
|
|
KIAA1598
|
KIAA1598
|
|
chr7_+_29486010
|
0.103
|
NM_001039936
|
CHN2
|
chimerin (chimaerin) 2
|
|
chr1_+_24713396
|
0.103
|
|
RCAN3
|
RCAN family member 3
|
|
chrX_-_10495476
|
0.101
|
NM_001193278
NM_001193279
NM_001193280
NM_001193281
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr6_-_32920689
|
0.101
|
NM_004159
|
PSMB8
|
proteasome (prosome, macropain) subunit, beta type, 8 (large multifunctional peptidase 7)
|
|
chr6_+_31731667
|
0.101
|
|
APOM
|
apolipoprotein M
|
|
chr8_-_17599438
|
0.101
|
NM_020749
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr19_-_38052481
|
0.101
|
NM_001126335
NM_014270
|
SLC7A9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9
|
|
chrM_+_13453
|
0.097
|
|
|
|
|
chr12_-_84754289
|
0.097
|
NM_005447
|
RASSF9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9
|
|
chrX_-_110541029
|
0.097
|
NM_000555
|
DCX
|
doublecortin
|
|
chr7_+_23252945
|
0.096
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr3_-_180452338
|
0.094
|
NM_171830
|
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3
|
|
chr16_-_70763849
|
0.093
|
NM_001160213
|
PMFBP1
|
polyamine modulated factor 1 binding protein 1
|
|
chr12_-_10853940
|
0.088
|
NM_023917
|
TAS2R9
|
taste receptor, type 2, member 9
|
|
chr10_-_116276651
|
0.088
|
NM_006720
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr13_+_52114565
|
0.085
|
|
HNRNPA1L2
|
heterogeneous nuclear ribonucleoprotein A1-like 2
|
|
chr14_-_20642629
|
0.085
|
NM_001102454
|
ZNF219
|
zinc finger protein 219
|
|
chrY_-_2715478
|
0.084
|
NM_003140
|
SRY
|
sex determining region Y
|
|
chr13_-_47885248
|
0.084
|
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr10_+_32896656
|
0.084
|
NM_024688
|
C10orf68
|
chromosome 10 open reading frame 68
|
|
chr3_+_142175329
|
0.084
|
|
SLC25A36
|
solute carrier family 25, member 36
|
|
chr21_+_34112456
|
0.082
|
|
ITSN1
|
intersectin 1 (SH3 domain protein)
|
|
chr2_+_223433919
|
0.082
|
NM_004457
NM_203372
|
ACSL3
|
acyl-CoA synthetase long-chain family member 3
|
|
chr6_+_31731647
|
0.082
|
NM_019101
|
APOM
|
apolipoprotein M
|
|
chr7_+_23252802
|
0.081
|
NM_001005340
NM_002510
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr1_-_151296611
|
0.081
|
NM_005988
|
SPRR2A
|
small proline-rich protein 2A
|
|
chr12_-_23995109
|
0.080
|
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr9_+_124835608
|
0.079
|
|
RABGAP1
GPR21
|
RAB GTPase activating protein 1
G protein-coupled receptor 21
|
|
chr15_+_57696200
|
0.077
|
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type
|
|
chr6_-_136829705
|
0.077
|
NM_001198615
|
MAP7
|
microtubule-associated protein 7
|
|
chr7_+_23252909
|
0.076
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr10_-_11614279
|
0.075
|
NM_001080491
|
USP6NL
|
USP6 N-terminal like
|
|
chr6_+_29187565
|
0.072
|
NM_001005216
|
OR2J3
|
olfactory receptor, family 2, subfamily J, member 3
|
|
chrM_-_15569
|
0.070
|
|
|
|
|
chr7_-_24764082
|
0.069
|
NM_001127454
NM_004403
|
DFNA5
|
deafness, autosomal dominant 5
|
|
chr15_-_63199228
|
0.069
|
|
PDCD7
|
programmed cell death 7
|
|
chr17_-_8364942
|
0.068
|
|
MYH10
|
myosin, heavy chain 10, non-muscle
|
|
chr12_+_117067214
|
0.068
|
|
PEBP1
|
phosphatidylethanolamine binding protein 1
|
|
chr12_-_8706637
|
0.067
|
NM_003480
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr2_-_74472486
|
0.067
|
|
DCTN1
|
dynactin 1
|
|
chr2_-_222867171
|
0.066
|
|
PAX3
|
paired box 3
|
|
chr19_-_60458915
|
0.066
|
|
PPP6R1
|
protein phosphatase 6, regulatory subunit 1
|
|
chr7_-_22518200
|
0.065
|
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1
|
|
chr1_-_158759606
|
0.065
|
NM_001184714
NM_001184715
NM_001184716
NM_052931
|
SLAMF6
|
SLAM family member 6
|
|
chr12_-_23628774
|
0.064
|
NM_178010
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr5_+_92944680
|
0.063
|
NM_005654
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr20_-_33783409
|
0.063
|
|
RBM39
|
RNA binding motif protein 39
|
|
chr2_-_74472480
|
0.063
|
|
DCTN1
|
dynactin 1
|
|
chrM_+_4465
|
0.061
|
|
|
|
|
chr13_-_47885018
|
0.060
|
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr16_-_28528780
|
0.060
|
NM_001055
NM_177529
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1
|
|
chr1_+_170894780
|
0.060
|
NM_000639
|
FASLG
|
Fas ligand (TNF superfamily, member 6)
|
|
chr1_+_169747788
|
0.060
|
|
PRRC2C
|
proline-rich coiled-coil 2C
|
|
chr6_-_11490487
|
0.060
|
NM_001142393
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chrX_-_106248674
|
0.059
|
|
RBM41
|
RNA binding motif protein 41
|
|
chr2_-_74472428
|
0.056
|
|
DCTN1
|
dynactin 1
|
|
chr14_-_106282128
|
0.055
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr12_-_48902692
|
0.055
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr3_+_44891101
|
0.054
|
NM_003241
|
TGM4
|
transglutaminase 4 (prostate)
|
|
chr3_+_100840129
|
0.053
|
NM_001850
NM_020351
|
COL8A1
|
collagen, type VIII, alpha 1
|
|
chr9_-_103187082
|
0.053
|
NM_001701
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase)
|
|
chr7_+_141110365
|
0.051
|
NM_016943
|
TAS2R3
|
taste receptor, type 2, member 3
|
|
chr7_+_65057942
|
0.050
|
|
VKORC1L1
|
vitamin K epoxide reductase complex, subunit 1-like 1
|
|
chr1_-_13263334
|
0.050
|
NM_001012276
|
PRAMEF8
PRAMEF7
|
PRAME family member 8
PRAME family member 7
|
|
chr22_-_41372920
|
0.049
|
NM_001129819
NM_001171661
NM_007326
|
CYB5R3
|
cytochrome b5 reductase 3
|
|
chr15_-_50270856
|
0.049
|
NM_016194
|
GNB5
|
guanine nucleotide binding protein (G protein), beta 5
|
|
chr20_+_1041905
|
0.049
|
NM_178578
|
PSMF1
|
proteasome (prosome, macropain) inhibitor subunit 1 (PI31)
|
|
chr5_+_36644187
|
0.049
|
NM_001166695
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr14_+_63750611
|
0.049
|
NM_182913
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2
|
|
chr11_-_73559507
|
0.049
|
NM_015531
|
C2CD3
|
C2 calcium-dependent domain containing 3
|
|
chr15_+_22652790
|
0.047
|
NM_022805
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N
|
|
chr8_+_92184022
|
0.046
|
NM_001129890
|
LRRC69
|
leucine rich repeat containing 69
|
|
chr15_+_69220841
|
0.046
|
NM_024817
|
THSD4
|
thrombospondin, type I, domain containing 4
|
|
chr8_+_67096344
|
0.046
|
NM_033105
|
DNAJC5B
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta
|
|
chr1_-_115039614
|
0.045
|
NM_000036
NM_001172626
|
AMPD1
|
adenosine monophosphate deaminase 1
|
|
chr8_+_22909305
|
0.045
|
NM_001160037
|
RHOBTB2
|
Rho-related BTB domain containing 2
|
|
chr1_+_234748187
|
0.045
|
NM_201545
|
LGALS8
|
lectin, galactoside-binding, soluble, 8
|
|
chr16_-_70763524
|
0.045
|
NM_031293
|
PMFBP1
|
polyamine modulated factor 1 binding protein 1
|
|
chr9_+_116943917
|
0.044
|
NM_017418
|
DEC1
|
deleted in esophageal cancer 1
|
|
chr20_+_34163671
|
0.044
|
NM_177996
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr11_-_73856241
|
0.043
|
NM_005472
|
KCNE3
|
potassium voltage-gated channel, Isk-related family, member 3
|