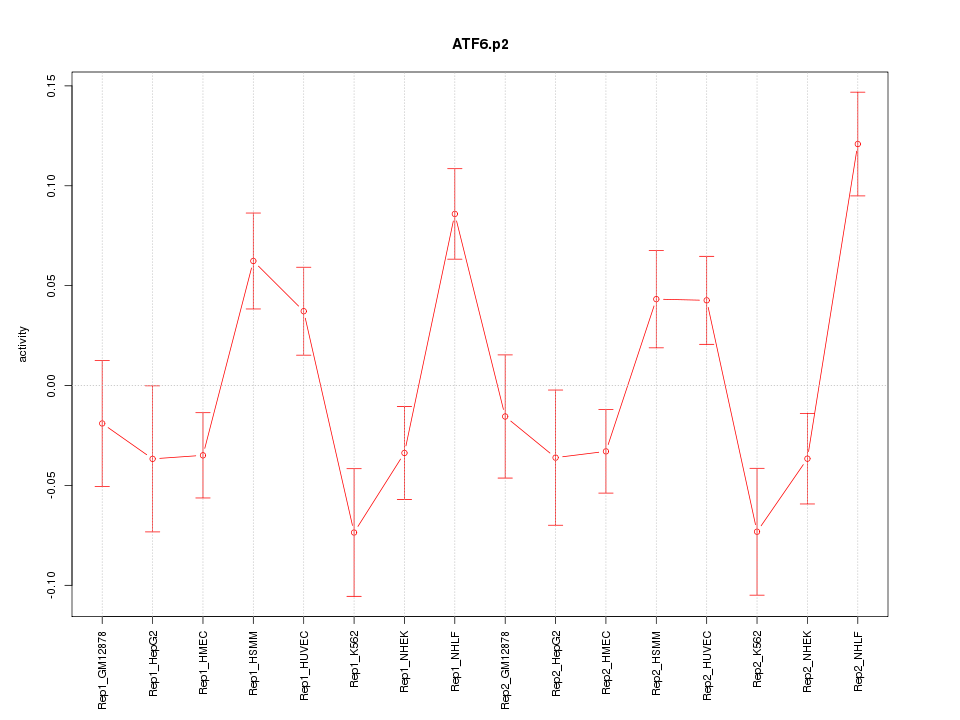
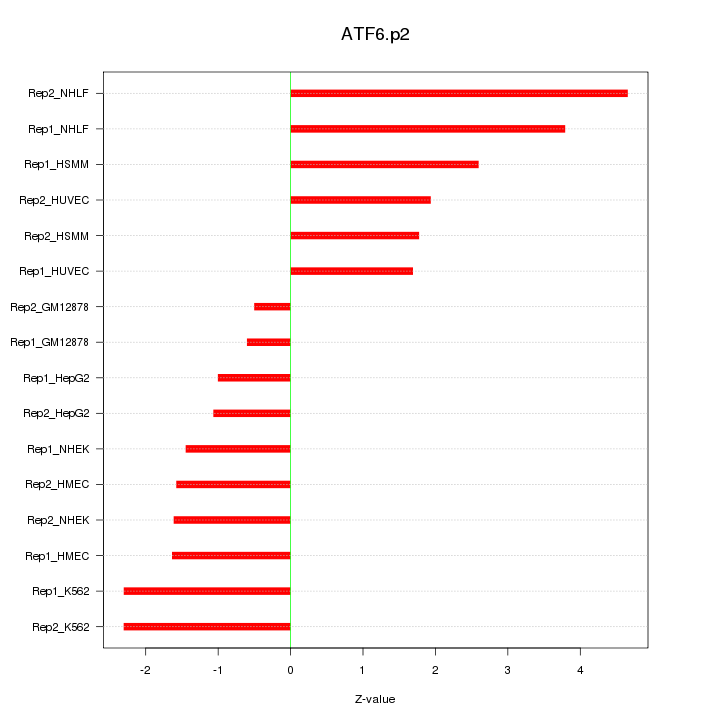
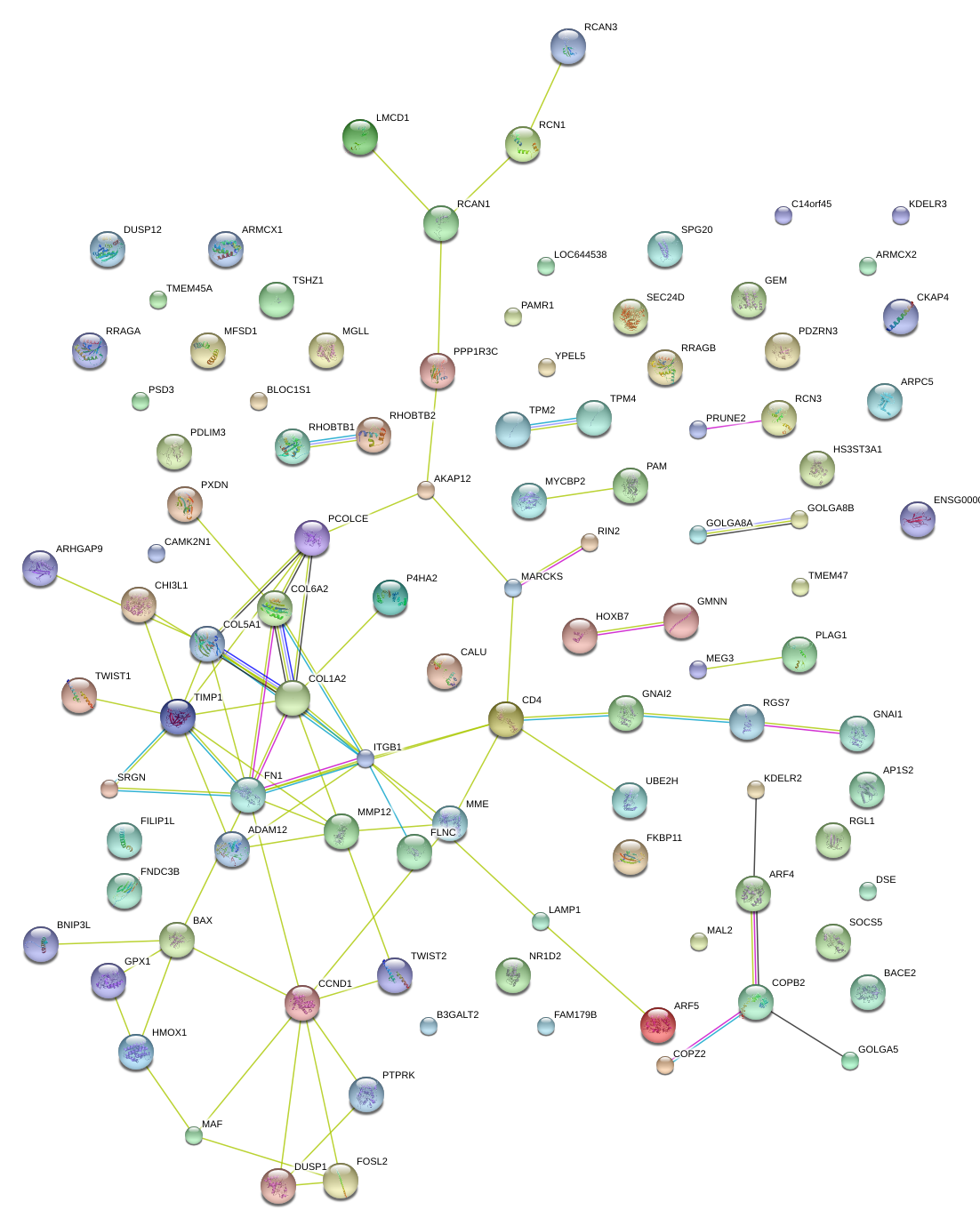
|
chrX_-_100801472
|
6.276
|
NM_014782
NM_177949
|
ARMCX2
|
armadillo repeat containing, X-linked 2
|
|
chr6_+_151602787
|
4.485
|
NM_005100
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr7_+_93861808
|
4.151
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr17_-_44043361
|
3.676
|
NM_004502
|
HOXB7
|
homeobox B7
|
|
chrX_-_34585287
|
3.602
|
NM_031442
|
TMEM47
|
transmembrane protein 47
|
|
chr19_+_54723044
|
3.523
|
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr17_-_44042924
|
3.359
|
|
HOXB7
|
homeobox B7
|
|
chr19_+_16048266
|
3.247
|
|
TPM4
|
tropomyosin 4
|
|
chr19_+_16048315
|
3.112
|
|
TPM4
|
tropomyosin 4
|
|
chrX_+_100692071
|
3.071
|
NM_016608
|
ARMCX1
|
armadillo repeat containing, X-linked 1
|
|
chr19_+_16048306
|
3.006
|
|
TPM4
|
tropomyosin 4
|
|
chr11_-_35503720
|
2.741
|
NM_001001991
NM_015430
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr12_-_105165805
|
2.699
|
NM_006825
|
CKAP4
|
cytoskeleton-associated protein 4
|
|
chr1_+_181871830
|
2.698
|
NM_015149
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr2_-_1727292
|
2.498
|
NM_012293
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr21_+_46369606
|
2.450
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr19_+_16048047
|
2.371
|
NM_003290
|
TPM4
|
tropomyosin 4
|
|
chr20_+_19863716
|
2.300
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr3_-_73756668
|
2.293
|
NM_015009
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr7_+_128257698
|
2.274
|
NM_001127487
NM_001458
|
FLNC
|
filamin C, gamma
|
|
chr22_+_37193961
|
2.197
|
NM_006855
NM_016657
|
KDELR3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3
|
|
chr2_-_1727247
|
2.161
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr5_+_102229612
|
2.152
|
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr3_-_57558193
|
2.150
|
NM_001660
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr3_-_49370755
|
2.101
|
NM_000581
NM_201397
|
GPX1
|
glutathione peroxidase 1
|
|
chr6_+_1336077
|
2.085
|
|
|
|
|
chr3_-_49370682
|
2.073
|
|
GPX1
|
glutathione peroxidase 1
|
|
chr17_-_44043278
|
2.047
|
|
HOXB7
|
homeobox B7
|
|
chr5_+_102229425
|
2.040
|
NM_000919
NM_001177306
NM_138766
NM_138821
NM_138822
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr5_-_172130766
|
1.977
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr14_+_44501083
|
1.944
|
NM_015091
|
FAM179B
|
family with sequence similarity 179, member B
|
|
chr3_+_160002582
|
1.911
|
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chr19_+_54149941
|
1.901
|
|
BAX
|
BCL2-associated X protein
|
|
chr10_-_62373828
|
1.897
|
NM_014836
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chr8_+_26296439
|
1.874
|
NM_004331
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like
|
|
chr3_-_57558130
|
1.864
|
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr19_+_54149843
|
1.848
|
NM_004324
NM_138761
NM_138763
NM_138764
|
BAX
|
BCL2-associated X protein
|
|
chr11_+_69165119
|
1.847
|
|
CCND1
|
cyclin D1
|
|
chr3_+_160002605
|
1.805
|
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chr1_-_191422346
|
1.801
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr11_+_69165018
|
1.790
|
NM_053056
|
CCND1
|
cyclin D1
|
|
chr22_+_34107087
|
1.774
|
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chrX_-_15782859
|
1.770
|
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr7_-_19123765
|
1.756
|
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr9_-_78710698
|
1.746
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr2_-_1727324
|
1.740
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr3_-_57558120
|
1.732
|
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr2_+_46779553
|
1.716
|
NM_014011
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr19_+_54149963
|
1.691
|
|
BAX
|
BCL2-associated X protein
|
|
chr22_+_34107051
|
1.679
|
NM_002133
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr7_-_19123787
|
1.671
|
NM_000474
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr2_+_46779827
|
1.667
|
NM_144949
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr8_-_95343716
|
1.652
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chrX_-_15782803
|
1.648
|
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr3_-_57557906
|
1.647
|
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr7_+_93862137
|
1.609
|
|
COL1A2
PLAG1
|
collagen, type I, alpha 2
pleiomorphic adenoma gene 1
|
|
chr12_+_54396099
|
1.552
|
|
BLOC1S1-RDH5
BLOC1S1
|
BLOC1S1-RDH5 read-through transcript
biogenesis of lysosomal organelles complex-1, subunit 1
|
|
chrX_+_47326594
|
1.545
|
NM_003254
|
TIMP1
|
TIMP metallopeptidase inhibitor 1
|
|
chr10_-_128067062
|
1.523
|
NM_003474
NM_021641
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr7_+_128166581
|
1.518
|
NM_001130674
NM_001219
|
CALU
|
calumenin
|
|
chr3_+_160002619
|
1.512
|
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chr12_-_47604930
|
1.489
|
NM_001143781
|
FKBP11
|
FK506 binding protein 11, 19 kDa
|
|
chr11_+_32068990
|
1.450
|
NM_002901
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain
|
|
chrX_+_47326746
|
1.433
|
|
TIMP1
|
TIMP metallopeptidase inhibitor 1
|
|
chr13_+_112999538
|
1.429
|
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr9_+_136673464
|
1.426
|
NM_000093
|
COL5A1
|
collagen, type V, alpha 1
|
|
chrX_-_15782842
|
1.425
|
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chrX_-_15782826
|
1.423
|
|
|
|
|
chr17_-_43470134
|
1.420
|
NM_016429
|
COPZ2
|
coatomer protein complex, subunit zeta 2
|
|
chr3_+_156280772
|
1.408
|
NM_007289
|
MME
|
membrane metallo-endopeptidase
|
|
chr2_-_216009015
|
1.396
|
NM_002026
NM_054034
NM_212474
NM_212476
NM_212478
NM_212482
|
FN1
|
fibronectin 1
|
|
chr3_+_50259329
|
1.383
|
NM_001166425
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2
|
|
chr3_+_160002408
|
1.381
|
NM_001167903
NM_022736
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chr1_-_20685314
|
1.375
|
NM_018584
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr1_-_181871508
|
1.351
|
NM_005717
|
ARPC5
|
actin related protein 2/3 complex, subunit 5, 16kDa
|
|
chr6_+_116798791
|
1.341
|
NM_013352
|
DSE
|
dermatan sulfate epimerase
|
|
chr7_-_6490214
|
1.338
|
NM_001100603
NM_006854
|
KDELR2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2
|
|
chr4_-_186693574
|
1.332
|
NM_001114107
NM_014476
|
PDLIM3
|
PDZ and LIM domain 3
|
|
chr7_-_129379947
|
1.330
|
NM_003344
NM_182697
|
UBE2H
|
ubiquitin-conjugating enzyme E2H (UBC8 homolog, yeast)
|
|
chr3_+_173240126
|
1.323
|
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr1_-_181871447
|
1.310
|
|
ARPC5
|
actin related protein 2/3 complex, subunit 5, 16kDa
|
|
chr14_+_100362204
|
1.310
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chrX_-_15783015
|
1.308
|
NM_003916
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr18_+_71051713
|
1.306
|
NM_005786
|
TSHZ1
|
teashirt zinc finger homeobox 1
|
|
chr2_+_28469199
|
1.305
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr1_-_181871490
|
1.299
|
|
ARPC5
|
actin related protein 2/3 complex, subunit 5, 16kDa
|
|
chr6_-_128883125
|
1.287
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr12_+_54396118
|
1.286
|
|
BLOC1S1
|
biogenesis of lysosomal organelles complex-1, subunit 1
|
|
chr3_+_8518508
|
1.277
|
NM_014583
|
LMCD1
|
LIM and cysteine-rich domains 1
|
|
chr2_+_30223253
|
1.277
|
NM_001127399
NM_001127400
NM_001127401
NM_016061
|
YPEL5
|
yippee-like 5 (Drosophila)
|
|
chr1_-_181871462
|
1.265
|
|
ARPC5
|
actin related protein 2/3 complex, subunit 5, 16kDa
|
|
chr13_-_35818777
|
1.249
|
NM_001142295
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr7_-_6490375
|
1.246
|
|
KDELR2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2
|
|
chr4_-_119976631
|
1.233
|
|
SEC24D
|
SEC24 family, member D (S. cerevisiae)
|
|
chrX_-_15782792
|
1.228
|
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr3_+_23962518
|
1.226
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr1_-_181871473
|
1.225
|
|
ARPC5
|
actin related protein 2/3 complex, subunit 5, 16kDa
|
|
chr7_+_100041143
|
1.221
|
|
PCOLCE
|
procollagen C-endopeptidase enhancer
|
|
chr4_-_119976736
|
1.215
|
NM_014822
|
SEC24D
|
SEC24 family, member D (S. cerevisiae)
|
|
chr3_+_101694098
|
1.212
|
NM_018004
|
TMEM45A
|
transmembrane protein 45A
|
|
chr12_+_54396086
|
1.210
|
NM_001487
|
BLOC1S1
|
biogenesis of lysosomal organelles complex-1, subunit 1
|
|
chr8_-_18585721
|
1.200
|
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr13_-_35818491
|
1.199
|
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr7_+_128166631
|
1.199
|
|
CALU
|
calumenin
|
|
chr3_+_173240107
|
1.197
|
NM_022763
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr3_+_101694244
|
1.195
|
|
TMEM45A
|
transmembrane protein 45A
|
|
chr1_-_181871416
|
1.178
|
|
ARPC5
|
actin related protein 2/3 complex, subunit 5, 16kDa
|
|
chr3_+_160002571
|
1.177
|
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chrX_+_133952633
|
1.174
|
NM_001163438
|
LOC644538
|
hypothetical protein LOC644538
|
|
chr3_-_140591201
|
1.167
|
NM_004766
|
COPB2
|
coatomer protein complex, subunit beta 2 (beta prime)
|
|
chr8_-_95343696
|
1.164
|
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr1_-_239587007
|
1.161
|
|
RGS7
|
regulator of G-protein signaling 7
|
|
chr7_+_128166664
|
1.157
|
|
CALU
|
calumenin
|
|
chr21_+_41461597
|
1.155
|
NM_012105
NM_138991
NM_138992
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chr14_+_92330383
|
1.152
|
NM_005113
|
GOLGA5
|
golgin A5
|
|
chr8_+_120289735
|
1.151
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr7_+_128166636
|
1.145
|
|
CALU
|
calumenin
|
|
chr21_+_46370220
|
1.142
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr10_+_70517823
|
1.142
|
NM_002727
|
SRGN
|
serglycin
|
|
chr3_-_101077605
|
1.138
|
NM_014890
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr1_-_201420927
|
1.132
|
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chr14_+_73555802
|
1.121
|
NM_025057
|
C14orf45
|
chromosome 14 open reading frame 45
|
|
chr3_-_129023850
|
1.115
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr10_-_93382837
|
1.110
|
NM_005398
|
PPP1R3C
|
protein phosphatase 1, regulatory (inhibitor) subunit 3C
|
|
chr16_-_78192111
|
1.106
|
NM_001031804
NM_005360
|
MAF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian)
|
|
chr13_-_35818603
|
1.097
|
NM_001142296
NM_015087
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr10_-_33287137
|
1.094
|
|
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12)
|
|
chr1_-_239587090
|
1.090
|
NM_002924
|
RGS7
|
regulator of G-protein signaling 7
|
|
chr14_+_100362236
|
1.086
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr3_-_140591111
|
1.083
|
|
COPB2
|
coatomer protein complex, subunit beta 2 (beta prime)
|
|
chr6_+_114285209
|
1.078
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr5_-_131591380
|
1.069
|
NM_001017974
NM_001142598
NM_001142599
|
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II
|
|
chr9_-_35679902
|
1.048
|
NM_003289
NM_213674
|
TPM2
|
tropomyosin 2 (beta)
|
|
chr10_-_33287184
|
1.029
|
NM_002211
|
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12)
|
|
chr1_+_182040837
|
1.029
|
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr17_-_13445942
|
1.029
|
NM_006042
|
HS3ST3A1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1
|
|
chr2_+_30223304
|
1.010
|
|
YPEL5
|
yippee-like 5 (Drosophila)
|
|
chr12_+_6768934
|
1.006
|
NM_001195014
NM_001195015
NM_001195016
NM_001195017
|
CD4
|
CD4 molecule
|
|
chr19_+_54722686
|
1.002
|
NM_020650
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chrX_+_55760774
|
1.002
|
NM_006064
NM_016656
|
RRAGB
|
Ras-related GTP binding B
|
|
chr3_-_188946168
|
0.995
|
NM_001706
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr7_+_128166688
|
0.992
|
|
CALU
|
calumenin
|
|
chr19_+_50041400
|
0.988
|
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr19_-_47155340
|
0.983
|
NM_006423
|
RABAC1
|
Rab acceptor 1 (prenylated)
|
|
chr18_+_7557313
|
0.982
|
NM_001105244
NM_002845
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chr14_-_38642165
|
0.979
|
NM_006364
|
SEC23A
|
Sec23 homolog A (S. cerevisiae)
|
|
chr16_+_31390567
|
0.968
|
NM_001042454
NM_001164719
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr10_-_111673191
|
0.959
|
|
XPNPEP1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble
|
|
chrX_-_100759562
|
0.955
|
NM_001009584
NM_001184768
NM_019007
|
ARMCX6
|
armadillo repeat containing, X-linked 6
|
|
chr1_+_99502435
|
0.955
|
NM_001166252
NM_014839
|
LPPR4
|
lipid phosphate phosphatase-related protein type 4
|
|
chr1_+_148388719
|
0.953
|
NM_016274
|
PLEKHO1
|
pleckstrin homology domain containing, family O member 1
|
|
chr7_+_65307816
|
0.947
|
|
TPST1
|
tyrosylprotein sulfotransferase 1
|
|
chr14_-_22891865
|
0.947
|
NM_016609
|
SLC22A17
|
solute carrier family 22, member 17
|
|
chrX_-_128485134
|
0.940
|
NM_003069
NM_139035
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chr2_+_30223372
|
0.937
|
|
YPEL5
|
yippee-like 5 (Drosophila)
|
|
chr17_-_76064842
|
0.936
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr20_+_34523231
|
0.933
|
NM_001042486
|
DLGAP4
|
discs, large (Drosophila) homolog-associated protein 4
|
|
chr7_+_65307691
|
0.932
|
NM_003596
|
TPST1
|
tyrosylprotein sulfotransferase 1
|
|
chr12_+_6703686
|
0.909
|
NM_001164093
|
COPS7A
|
COP9 constitutive photomorphogenic homolog subunit 7A (Arabidopsis)
|
|
chr3_+_130641828
|
0.908
|
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas)
|
|
chr3_+_130641779
|
0.906
|
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas)
|
|
chr19_-_11166186
|
0.906
|
NM_015493
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chr15_-_38000379
|
0.898
|
NM_007223
|
GPR176
|
G protein-coupled receptor 176
|
|
chr3_+_130641639
|
0.897
|
NM_018262
NM_052985
NM_052989
NM_052990
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas)
|
|
chr17_-_37828643
|
0.897
|
NM_012232
|
PTRF
|
polymerase I and transcript release factor
|
|
chr3_-_109423858
|
0.894
|
NM_018010
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas)
|
|
chr1_+_99502655
|
0.894
|
|
LPPR4
|
lipid phosphate phosphatase-related protein type 4
|
|
chr7_-_37922902
|
0.891
|
|
SFRP4
|
secreted frizzled-related protein 4
|
|
chr3_+_130641840
|
0.889
|
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas)
|
|
chr16_+_15435825
|
0.886
|
NM_033201
|
C16orf45
|
chromosome 16 open reading frame 45
|
|
chr3_+_8518539
|
0.882
|
|
LMCD1
|
LIM and cysteine-rich domains 1
|
|
chr8_+_133856793
|
0.879
|
|
PHF20L1
|
PHD finger protein 20-like 1
|
|
chr3_+_130641765
|
0.875
|
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas)
|
|
chr16_+_31027255
|
0.875
|
|
BCKDK
|
branched chain ketoacid dehydrogenase kinase
|
|
chr6_-_90119158
|
0.866
|
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1 (UBC6 homolog, yeast)
|
|
chr16_+_31027231
|
0.866
|
|
BCKDK
|
branched chain ketoacid dehydrogenase kinase
|
|
chrX_-_128485121
|
0.864
|
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chr14_-_38642029
|
0.861
|
|
SEC23A
|
Sec23 homolog A (S. cerevisiae)
|
|
chr1_+_158579685
|
0.857
|
NM_015331
|
NCSTN
|
nicastrin
|
|
chr11_+_69165159
|
0.855
|
|
CCND1
|
cyclin D1
|
|
chr10_-_118022965
|
0.852
|
NM_005264
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr14_+_61231795
|
0.851
|
NM_001530
NM_181054
|
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor)
|
|
chr21_+_37367432
|
0.843
|
NM_001001894
|
TTC3
|
tetratricopeptide repeat domain 3
|
|
chr1_+_158579796
|
0.840
|
|
NCSTN
|
nicastrin
|
|
chr11_+_46255737
|
0.837
|
NM_052854
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1
|
|
chrX_+_102498035
|
0.835
|
NM_001006612
NM_001006613
NM_001006614
NM_016303
|
WBP5
|
WW domain binding protein 5
|
|
chr12_+_6768866
|
0.829
|
NM_000616
|
CD4
|
CD4 molecule
|
|
chr12_+_6768966
|
0.823
|
|
CD4
|
CD4 molecule
|
|
chr6_-_90119068
|
0.822
|
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1 (UBC6 homolog, yeast)
|
|
chr3_+_150191884
|
0.820
|
NM_001184720
NM_001184721
NM_004130
|
GYG1
|
glycogenin 1
|
|
chr13_+_112999590
|
0.815
|
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr7_-_27191287
|
0.809
|
NM_005523
|
HOXA11
|
homeobox A11
|
|
chr16_+_31027249
|
0.809
|
|
BCKDK
|
branched chain ketoacid dehydrogenase kinase
|
|
chr11_+_32069183
|
0.808
|
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain
|
|
chr2_+_241903395
|
0.806
|
NM_001008491
NM_006155
|
SEPT2
|
septin 2
|
|
chr4_-_54625223
|
0.800
|
|
CHIC2
|
cysteine-rich hydrophobic domain 2
|
|
chr17_+_30938433
|
0.799
|
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit
|
|
chr2_-_86704446
|
0.794
|
|
RNF103
|
ring finger protein 103
|
|
chr3_+_62279707
|
0.791
|
|
C3orf14
|
chromosome 3 open reading frame 14
|
|
chr4_-_54625528
|
0.787
|
NM_012110
|
CHIC2
|
cysteine-rich hydrophobic domain 2
|