|
chr22_-_21231421
|
1.865
|
NM_206955
NM_206956
NM_006115
NM_206953
NM_206954
|
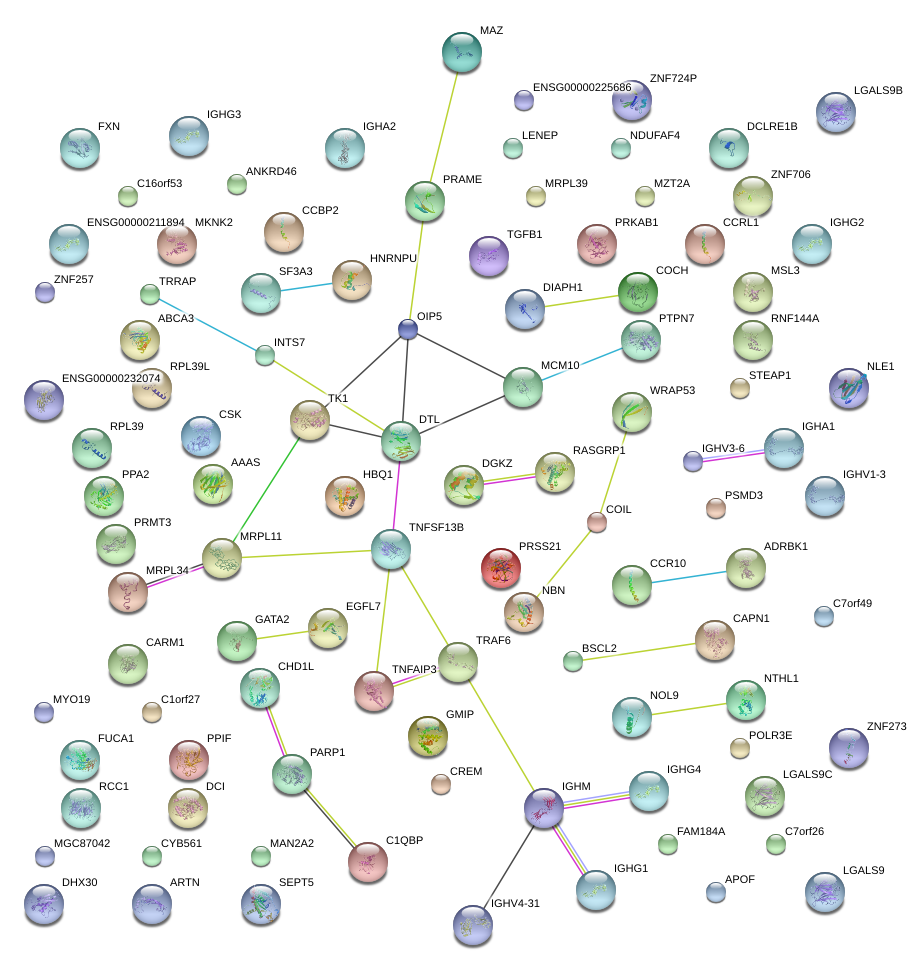
PRAME
|
preferentially expressed antigen in melanoma
|
|
chr22_+_21027538
|
1.259
|
|
|
|
|
chr1_+_210275541
|
1.103
|
NM_016448
|
DTL
|
denticleless homolog (Drosophila)
|
|
chr22_+_18085990
|
1.056
|
|
SEPT5-GP1BB
SEPT5
|
SEPT5-GP1BB read-through transcript
septin 5
|
|
chr22_+_21053972
|
1.019
|
|
|
|
|
chr1_-_224662156
|
1.008
|
|
PARP1
|
poly (ADP-ribose) polymerase 1
|
|
chr14_+_30413393
|
0.988
|
NM_001135058
NM_004086
|
COCH
|
coagulation factor C homolog, cochlin (Limulus polyphemus)
|
|
chr10_+_80777199
|
0.961
|
NM_005729
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr10_+_13243553
|
0.914
|
NM_018518
NM_182751
|
MCM10
|
minichromosome maintenance complex component 10
|
|
chr1_-_200396354
|
0.910
|
NM_002832
|
PTPN7
|
protein tyrosine phosphatase, non-receptor type 7
|
|
chr16_+_170334
|
0.910
|
NM_005331
|
HBQ1
|
hemoglobin, theta 1
|
|
chr10_+_80777248
|
0.901
|
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr17_-_5283104
|
0.880
|
|
C1QBP
|
complement component 1, q subcomponent binding protein
|
|
chr19_-_46551670
|
0.870
|
NM_000660
|
TGFB1
|
transforming growth factor, beta 1
|
|
chr16_-_2330738
|
0.855
|
NM_001089
|
ABCA3
|
ATP-binding cassette, sub-family A (ABC1), member 3
|
|
chr16_-_2330712
|
0.833
|
|
ABCA3
|
ATP-binding cassette, sub-family A (ABC1), member 3
|
|
chr16_+_2807228
|
0.831
|
|
PRSS21
|
protease, serine, 21 (testisin)
|
|
chr21_-_25901644
|
0.807
|
NM_017446
NM_080794
|
MRPL39
|
mitochondrial ribosomal protein L39
|
|
chr2_+_6974922
|
0.758
|
NM_014746
|
RNF144A
|
ring finger protein 144A
|
|
chr17_-_5283148
|
0.752
|
NM_001212
|
C1QBP
|
complement component 1, q subcomponent binding protein
|
|
chr22_+_21079355
|
0.745
|
|
|
|
|
chr15_+_72861767
|
0.738
|
|
CSK
|
c-src tyrosine kinase
|
|
chr17_-_30493254
|
0.729
|
|
NLE1
|
notchless homolog 1 (Drosophila)
|
|
chr3_-_188339882
|
0.708
|
NM_052969
|
RPL39L
|
ribosomal protein L39-like
|
|
chr1_+_114249549
|
0.688
|
|
DCLRE1B
|
DNA cross-link repair 1B
|
|
chr8_-_102286695
|
0.683
|
|
ZNF706
|
zinc finger protein 706
|
|
chr1_+_114249429
|
0.682
|
NM_022836
|
DCLRE1B
|
DNA cross-link repair 1B
|
|
chr8_-_102287044
|
0.669
|
NM_001042511
|
ZNF706
|
zinc finger protein 706
|
|
chr8_-_101641046
|
0.658
|
NM_198401
|
GAPDHP62
ANKRD46
|
glyceraldehyde 3 phosphate dehydrogenase pseudogene 62
ankyrin repeat domain 46
|
|
chr22_+_18085891
|
0.658
|
|
SEPT5
|
septin 5
|
|
chr1_-_224662307
|
0.657
|
|
PARP1
|
poly (ADP-ribose) polymerase 1
|
|
chr11_-_65962836
|
0.640
|
NM_016050
NM_170738
NM_170739
|
MRPL11
|
mitochondrial ribosomal protein L11
|
|
chr15_-_36644122
|
0.633
|
NM_001128602
NM_005739
|
RASGRP1
|
RAS guanyl releasing protein 1 (calcium and DAG-regulated)
|
|
chr1_-_24067347
|
0.632
|
NM_000147
|
FUCA1
|
fucosidase, alpha-L- 1, tissue
|
|
chr22_+_21116283
|
0.621
|
|
|
|
|
chr7_-_22506320
|
0.620
|
NM_001164460
NM_207342
|
MGC87042
|
STEAP family protein MGC87042
|
|
chr16_+_29735208
|
0.614
|
|
C16orf53
|
chromosome 16 open reading frame 53
|
|
chr10_+_80777234
|
0.611
|
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr8_+_43067817
|
0.609
|
|
|
|
|
chr10_+_35455986
|
0.597
|
|
CREM
|
cAMP responsive element modulator
|
|
chr1_-_224662368
|
0.590
|
|
PARP1
|
poly (ADP-ribose) polymerase 1
|
|
chr17_+_22996461
|
0.588
|
|
LGALS9
|
lectin, galactoside-binding, soluble, 9
|
|
chr12_+_118590039
|
0.576
|
NM_006253
|
PRKAB1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit
|
|
chr19_+_17277476
|
0.574
|
NM_023937
|
MRPL34
|
mitochondrial ribosomal protein L34
|
|
chr15_+_72861470
|
0.570
|
NM_001127190
NM_004383
|
CSK
|
c-src tyrosine kinase
|
|
chr8_-_102287083
|
0.570
|
NM_001042510
NM_016096
|
ZNF706
|
zinc finger protein 706
|
|
chr17_-_73694669
|
0.570
|
NM_003258
|
TK1
|
thymidine kinase 1, soluble
|
|
chr9_+_138680064
|
0.555
|
|
EGFL7
|
EGF-like-domain, multiple 7
|
|
chr17_-_52393364
|
0.555
|
NM_004645
|
COIL
|
coilin
|
|
chr7_+_6596183
|
0.551
|
NM_024067
|
C7orf26
|
chromosome 7 open reading frame 26
|
|
chr1_-_224662396
|
0.549
|
|
PARP1
|
poly (ADP-ribose) polymerase 1
|
|
chr11_+_20365788
|
0.549
|
|
PRMT3
|
protein arginine methyltransferase 3
|
|
chr3_-_129694717
|
0.545
|
NM_032638
|
GATA2
|
GATA binding protein 2
|
|
chr9_+_70840463
|
0.542
|
|
FXN
|
frataxin
|
|
chr7_-_134506070
|
0.542
|
NM_024033
|
C7orf49
|
chromosome 7 open reading frame 49
|
|
chr11_+_20365651
|
0.540
|
NM_001145166
NM_001145167
NM_005788
|
PRMT3
|
protein arginine methyltransferase 3
|
|
chr10_+_35455806
|
0.538
|
NM_001881
NM_182850
NM_182853
NM_183013
|
CREM
|
cAMP responsive element modulator
|
|
chr7_-_134506001
|
0.537
|
|
C7orf49
|
chromosome 7 open reading frame 49
|
|
chr1_-_224662419
|
0.537
|
NM_001618
|
PARP1
|
poly (ADP-ribose) polymerase 1
|
|
chr7_-_134506012
|
0.527
|
|
C7orf49
|
chromosome 7 open reading frame 49
|
|
chr7_+_63980305
|
0.526
|
|
ZNF273
|
zinc finger protein 273
|
|
chr22_+_20880112
|
0.521
|
|
|
|
|
chr3_+_47841478
|
0.513
|
|
DHX30
|
DEAH (Asp-Glu-Ala-His) box polypeptide 30
|
|
chr16_+_22216223
|
0.503
|
NM_018119
|
POLR3E
|
polymerase (RNA) III (DNA directed) polypeptide E (80kD)
|
|
chr10_+_80777265
|
0.502
|
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr7_-_134505923
|
0.498
|
|
C7orf49
|
chromosome 7 open reading frame 49
|
|
chr14_-_105644300
|
0.498
|
|
IGHV4-31
IGHA1
IGHA2
IGHG1
IGHG3
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
|
|
chr9_+_70840298
|
0.498
|
NM_000144
NM_001161706
NM_181425
|
FXN
|
frataxin
|
|
chr17_+_7532391
|
0.490
|
NM_001143992
NM_018081
|
WRAP53
|
WD repeat containing, antisense to TP53
|
|
chr1_+_28717234
|
0.489
|
NM_001048194
NM_001048195
NM_001269
|
RCC1
|
regulator of chromosome condensation 1
|
|
chr17_-_30493382
|
0.486
|
NM_001014445
NM_018096
|
NLE1
|
notchless homolog 1 (Drosophila)
|
|
chr8_-_91066060
|
0.484
|
NM_002485
|
NBN
|
nibrin
|
|
chr19_-_2002222
|
0.482
|
NM_017572
NM_199054
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chr16_-_2241561
|
0.479
|
NM_001178029
NM_001919
|
DCI
|
dodecenoyl-CoA isomerase
|
|
chr16_-_2037819
|
0.478
|
NM_002528
|
NTHL1
|
nth endonuclease III-like 1 (E. coli)
|
|
chr1_-_224662413
|
0.477
|
|
PARP1
|
poly (ADP-ribose) polymerase 1
|
|
chr4_-_106614562
|
0.475
|
NM_006903
NM_176866
NM_176867
NM_176869
|
PPA2
|
pyrophosphatase (inorganic) 2
|
|
chr1_-_38228179
|
0.463
|
NM_006802
|
SF3A3
|
splicing factor 3a, subunit 3, 60kDa
|
|
chr19_+_10843244
|
0.455
|
NM_199141
|
CARM1
|
coactivator-associated arginine methyltransferase 1
|
|
chr1_-_243093833
|
0.453
|
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A)
|
|
chr8_-_91065961
|
0.443
|
|
NBN
|
nibrin
|
|
chr11_-_62218759
|
0.442
|
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr16_-_2330594
|
0.441
|
|
ABCA3
|
ATP-binding cassette, sub-family A (ABC1), member 3
|
|
chr16_+_29725597
|
0.439
|
|
|
|
|
chr5_-_140978751
|
0.436
|
NM_001079812
NM_005219
|
DIAPH1
|
diaphanous homolog 1 (Drosophila)
|
|
chr2_-_131966416
|
0.435
|
|
MZT2A
|
mitotic spindle organizing protein 2A
|
|
chr16_+_29734785
|
0.433
|
|
C16orf53
|
chromosome 16 open reading frame 53
|
|
chr17_-_38087366
|
0.415
|
NM_016602
|
CCR10
|
chemokine (C-C motif) receptor 10
|
|
chr1_+_145180914
|
0.413
|
NM_004284
|
CHD1L
|
chromodomain helicase DNA binding protein 1-like
|
|
chr1_+_145180959
|
0.403
|
|
CHD1L
|
chromodomain helicase DNA binding protein 1-like
|
|
chr17_+_35390711
|
0.399
|
|
PSMD3
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 3
|
|
chr1_+_28717349
|
0.398
|
|
RCC1
|
regulator of chromosome condensation 1
|
|
chr12_-_52001610
|
0.398
|
NM_001173466
NM_015665
|
AAAS
|
achalasia, adrenocortical insufficiency, alacrimia
|
|
chr1_-_6537171
|
0.397
|
NM_024654
|
NOL9
|
nucleolar protein 9
|
|
chr15_+_89246780
|
0.396
|
|
MAN2A2
|
mannosidase, alpha, class 2A, member 2
|
|
chr1_-_200396328
|
0.389
|
|
PTPN7
|
protein tyrosine phosphatase, non-receptor type 7
|
|
chr17_-_31964723
|
0.389
|
|
MYO19
|
myosin XIX
|
|
chr6_-_97452416
|
0.389
|
|
NDUFAF4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 4
|
|
chr16_+_29725311
|
0.385
|
NM_001042539
NM_002383
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor)
|
|
chr11_+_64705261
|
0.383
|
NM_001198868
|
CAPN1
|
calpain 1, (mu/I) large subunit
|
|
chr1_-_210275402
|
0.380
|
NM_015434
|
INTS7
|
integrator complex subunit 7
|
|
chrX_+_11686629
|
0.377
|
NM_001193270
|
MSL3
|
male-specific lethal 3 homolog (Drosophila)
|
|
chr15_-_39412032
|
0.375
|
NM_007280
|
OIP5
|
Opa interacting protein 5
|
|
chr19_-_46550788
|
0.375
|
|
TGFB1
|
transforming growth factor, beta 1
|
|
chr7_+_98314020
|
0.368
|
NM_003496
|
TRRAP
|
transformation/transcription domain-associated protein
|
|
chr1_+_184611512
|
0.364
|
NM_001164245
NM_001164246
NM_017847
|
C1orf27
|
chromosome 1 open reading frame 27
|
|
chr11_+_66790517
|
0.360
|
|
ADRBK1
|
adrenergic, beta, receptor kinase 1
|
|
chr6_+_138230045
|
0.356
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr11_-_36488343
|
0.356
|
NM_004620
NM_145803
|
TRAF6
|
TNF receptor-associated factor 6
|
|
chr16_+_29734990
|
0.356
|
NM_024516
|
C16orf53
|
chromosome 16 open reading frame 53
|
|
chr6_-_119512035
|
0.354
|
NM_001100411
|
FAM184A
|
family with sequence similarity 184, member A
|
|
chr12_+_56406296
|
0.353
|
|
LOC100130776
|
hypothetical LOC100130776
|
|
chr12_-_52001515
|
0.352
|
|
AAAS
|
achalasia, adrenocortical insufficiency, alacrimia
|
|
chr19_-_19615452
|
0.348
|
NM_016573
|
GMIP
|
GEM interacting protein
|
|
chr17_-_58877276
|
0.347
|
NM_001017916
NM_001915
|
CYB561
|
cytochrome b-561
|
|
chr1_+_153232685
|
0.347
|
NM_018655
|
LENEP
|
lens epithelial protein
|
|
chr11_+_46325457
|
0.346
|
NM_003646
|
DGKZ
|
diacylglycerol kinase, zeta
|
|
chr7_-_38355949
|
0.345
|
|
|
|
|
chr19_+_10843369
|
0.344
|
|
CARM1
|
coactivator-associated arginine methyltransferase 1
|
|
chr19_-_53841064
|
0.344
|
NM_001217
|
CA11
|
carbonic anhydrase XI
|
|
chr16_-_65422365
|
0.341
|
NM_001018159
NM_001018160
NM_003905
|
NAE1
|
NEDD8 activating enzyme E1 subunit 1
|
|
chr22_+_20880159
|
0.339
|
|
IGLV6-57
|
immunoglobulin lambda variable 6-57
|
|
chr5_-_140978744
|
0.337
|
|
DIAPH1
|
diaphanous homolog 1 (Drosophila)
|
|
chr9_-_93751900
|
0.336
|
NM_004560
|
ROR2
|
receptor tyrosine kinase-like orphan receptor 2
|
|
chrX_+_11686198
|
0.335
|
NM_078628
NM_078629
|
MSL3
|
male-specific lethal 3 homolog (Drosophila)
|
|
chr1_+_95472298
|
0.335
|
NM_001128142
NM_015485
|
RWDD3
|
RWD domain containing 3
|
|
chr1_-_170679830
|
0.332
|
|
PIGC
|
phosphatidylinositol glycan anchor biosynthesis, class C
|
|
chr6_-_33028790
|
0.327
|
NM_006120
|
HLA-DMA
|
major histocompatibility complex, class II, DM alpha
|
|
chr8_-_74821610
|
0.326
|
|
STAU2
LOC729696
|
staufen, RNA binding protein, homolog 2 (Drosophila)
hypothetical protein LOC729696
|
|
chr11_+_66790740
|
0.326
|
|
ADRBK1
|
adrenergic, beta, receptor kinase 1
|
|
chr10_+_35456095
|
0.323
|
|
CREM
|
cAMP responsive element modulator
|
|
chr22_+_17800018
|
0.318
|
NM_003776
|
MRPL40
|
mitochondrial ribosomal protein L40
|
|
chr1_+_152511708
|
0.316
|
|
HAX1
|
HCLS1 associated protein X-1
|
|
chr1_-_94147574
|
0.315
|
NM_002061
|
GCLM
|
glutamate-cysteine ligase, modifier subunit
|
|
chr19_-_19615369
|
0.314
|
|
GMIP
|
GEM interacting protein
|
|
chr1_+_10413361
|
0.311
|
|
APITD1
|
apoptosis-inducing, TAF9-like domain 1
|
|
chr2_-_190893015
|
0.310
|
NM_014362
NM_198047
|
HIBCH
|
3-hydroxyisobutyryl-CoA hydrolase
|
|
chr10_+_92621221
|
0.310
|
|
RPP30
|
ribonuclease P/MRP 30kDa subunit
|
|
chr1_-_94147616
|
0.309
|
|
GCLM
|
glutamate-cysteine ligase, modifier subunit
|
|
chr5_+_132415674
|
0.309
|
|
HSPA4
|
heat shock 70kDa protein 4
|
|
chr12_-_119391918
|
0.309
|
NM_003769
|
SRSF9
|
serine/arginine-rich splicing factor 9
|
|
chr1_+_152511610
|
0.308
|
NM_001018837
NM_006118
|
HAX1
|
HCLS1 associated protein X-1
|
|
chr21_-_43400750
|
0.308
|
NM_001025203
NM_001025204
NM_006758
|
U2AF1
|
U2 small nuclear RNA auxiliary factor 1
|
|
chr1_-_94147537
|
0.306
|
|
GCLM
|
glutamate-cysteine ligase, modifier subunit
|
|
chr4_+_76077229
|
0.306
|
NM_015393
|
PARM1
|
prostate androgen-regulated mucin-like protein 1
|
|
chr11_+_118582231
|
0.305
|
|
CBL
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence
|
|
chr17_+_35390632
|
0.301
|
|
PSMD3
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 3
|
|
chr22_-_49311439
|
0.300
|
NM_001169110
|
SCO2
|
SCO cytochrome oxidase deficient homolog 2 (yeast)
|
|
chr5_+_157103280
|
0.300
|
NM_173491
|
LSM11
|
LSM11, U7 small nuclear RNA associated
|
|
chr1_+_16639822
|
0.300
|
|
NECAP2
|
NECAP endocytosis associated 2
|
|
chr9_-_129993615
|
0.297
|
NM_001131016
NM_001131017
NM_001131018
|
CIZ1
|
CDKN1A interacting zinc finger protein 1
|
|
chr14_-_106064887
|
0.297
|
|
LOC100126583
IGHV3-48
IGHA1
IGHM
|
hypothetical LOC100126583
immunoglobulin heavy variable 3-48
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant mu
|
|
chr21_-_43400672
|
0.297
|
|
U2AF1
|
U2 small nuclear RNA auxiliary factor 1
|
|
chr20_+_60311334
|
0.296
|
|
ADRM1
|
adhesion regulating molecule 1
|
|
chr13_+_32058610
|
0.294
|
|
PDS5B
|
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae)
|
|
chr14_-_105681377
|
0.293
|
|
IGHV4-31
IGHG1
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr17_+_35390579
|
0.292
|
NM_002809
|
PSMD3
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 3
|
|
chr6_-_159160386
|
0.291
|
|
EZR
|
ezrin
|
|
chr1_+_152511756
|
0.291
|
|
HAX1
|
HCLS1 associated protein X-1
|
|
chr14_-_105948734
|
0.291
|
|
LOC100126583
IGHV4-31
IGHA1
IGHA2
IGHG1
IGHG4
IGHM
|
hypothetical LOC100126583
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 4 (G4m marker)
immunoglobulin heavy constant mu
|
|
chr11_+_66790724
|
0.289
|
|
ADRBK1
|
adrenergic, beta, receptor kinase 1
|
|
chr17_+_35390607
|
0.289
|
|
PSMD3
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 3
|
|
chr16_-_730889
|
0.289
|
NM_022493
|
NARFL
|
nuclear prelamin A recognition factor-like
|
|
chr2_-_190892730
|
0.287
|
|
HIBCH
|
3-hydroxyisobutyryl-CoA hydrolase
|
|
chr12_-_54869568
|
0.286
|
NM_001130420
NM_003075
NM_139067
|
SMARCC2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2
|
|
chr22_+_21094094
|
0.286
|
|
|
|
|
chr17_-_77588844
|
0.285
|
NM_001195218
NM_016286
|
DCXR
|
dicarbonyl/L-xylulose reductase
|
|
chr6_+_41862397
|
0.284
|
|
TOMM6
|
translocase of outer mitochondrial membrane 6 homolog (yeast)
|
|
chr2_+_102456193
|
0.284
|
NM_001011552
|
SLC9A4
|
solute carrier family 9 (sodium/hydrogen exchanger), member 4
|
|
chr11_+_66790465
|
0.283
|
NM_001619
|
ADRBK1
|
adrenergic, beta, receptor kinase 1
|
|
chr13_+_32058651
|
0.282
|
|
PDS5B
|
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae)
|
|
chr6_-_159160387
|
0.278
|
|
EZR
|
ezrin
|
|
chr19_-_6432765
|
0.277
|
NM_024898
|
DENND1C
|
DENN/MADD domain containing 1C
|
|
chr17_+_45911216
|
0.275
|
|
RSAD1
|
radical S-adenosyl methionine domain containing 1
|
|
chr16_+_1143241
|
0.275
|
NM_001005407
NM_021098
|
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit
|
|
chr3_-_185218311
|
0.274
|
NM_001023587
NM_005688
|
ABCC5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5
|
|
chr17_-_18469569
|
0.274
|
|
CCDC144B
|
coiled-coil domain containing 144B
|
|
chr8_-_74821636
|
0.272
|
NM_001164380
NM_001164381
NM_001164382
NM_001164383
NM_001164385
NM_014393
|
STAU2
|
staufen, RNA binding protein, homolog 2 (Drosophila)
|
|
chr1_-_6684465
|
0.272
|
|
DNAJC11
|
DnaJ (Hsp40) homolog, subfamily C, member 11
|
|
chr16_-_88613287
|
0.271
|
NM_001042610
|
DBNDD1
|
dysbindin (dystrobrevin binding protein 1) domain containing 1
|
|
chr14_-_54438913
|
0.271
|
NM_000161
NM_001024024
NM_001024070
NM_001024071
|
GCH1
|
GTP cyclohydrolase 1
|
|
chr17_+_63144500
|
0.271
|
NM_015462
|
NOL11
|
nucleolar protein 11
|
|
chr16_-_65422304
|
0.269
|
|
NAE1
|
NEDD8 activating enzyme E1 subunit 1
|
|
chr18_+_42008063
|
0.266
|
|
C18orf25
|
chromosome 18 open reading frame 25
|
|
chr5_-_140978664
|
0.262
|
|
DIAPH1
|
diaphanous homolog 1 (Drosophila)
|
|
chr1_+_16639805
|
0.262
|
|
NECAP2
|
NECAP endocytosis associated 2
|
|
chr16_+_67902751
|
0.262
|
NM_013245
|
VPS4A
|
vacuolar protein sorting 4 homolog A (S. cerevisiae)
|
|
chr2_+_233927892
|
0.262
|
|
DGKD
|
diacylglycerol kinase, delta 130kDa
|
|
chr17_+_35390641
|
0.261
|
|
PSMD3
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 3
|
|
chr1_-_6684426
|
0.260
|
|
DNAJC11
|
DnaJ (Hsp40) homolog, subfamily C, member 11
|
|
chr19_+_43585668
|
0.259
|
|
FAM98C
|
family with sequence similarity 98, member C
|
|
chr11_+_66790505
|
0.259
|
|
ADRBK1
|
adrenergic, beta, receptor kinase 1
|
|
chr22_-_49311722
|
0.257
|
NM_001169109
|
SCO2
|
SCO cytochrome oxidase deficient homolog 2 (yeast)
|
|
chr20_+_60311443
|
0.257
|
|
ADRM1
|
adhesion regulating molecule 1
|
|
chr4_-_1980500
|
0.256
|
|
WHSC2
|
Wolf-Hirschhorn syndrome candidate 2
|
|
chr14_-_64359565
|
0.253
|
NM_000347
NM_001024858
|
SPTB
|
spectrin, beta, erythrocytic
|
|
chr15_+_48261649
|
0.253
|
NM_001159629
NM_003645
|
SLC27A2
|
solute carrier family 27 (fatty acid transporter), member 2
|
|
chr20_+_60311469
|
0.252
|
|
ADRM1
|
adhesion regulating molecule 1
|
|
chr1_-_168310232
|
0.252
|
NM_014970
|
KIFAP3
|
kinesin-associated protein 3
|
|
chr10_+_104144218
|
0.252
|
NM_001077493
NM_001077494
|
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100)
|