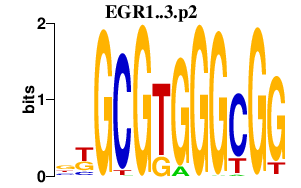
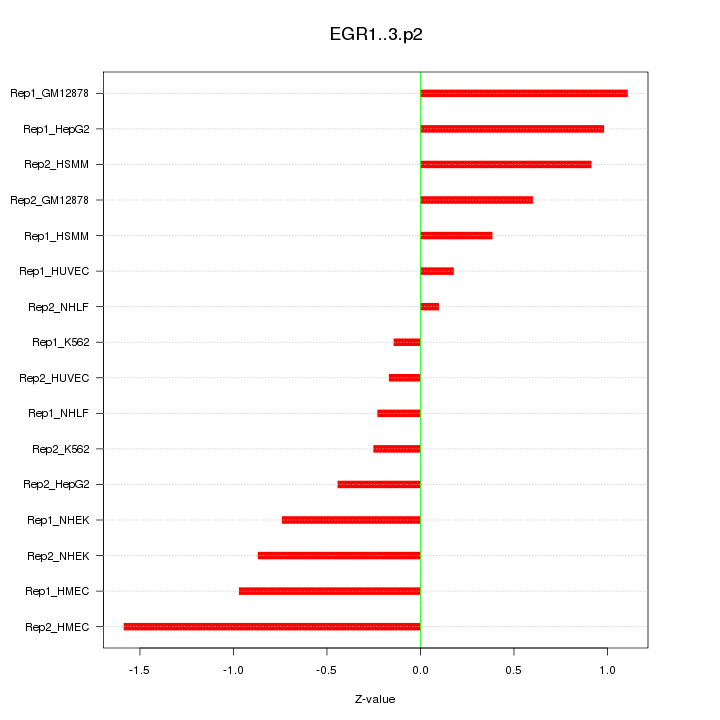
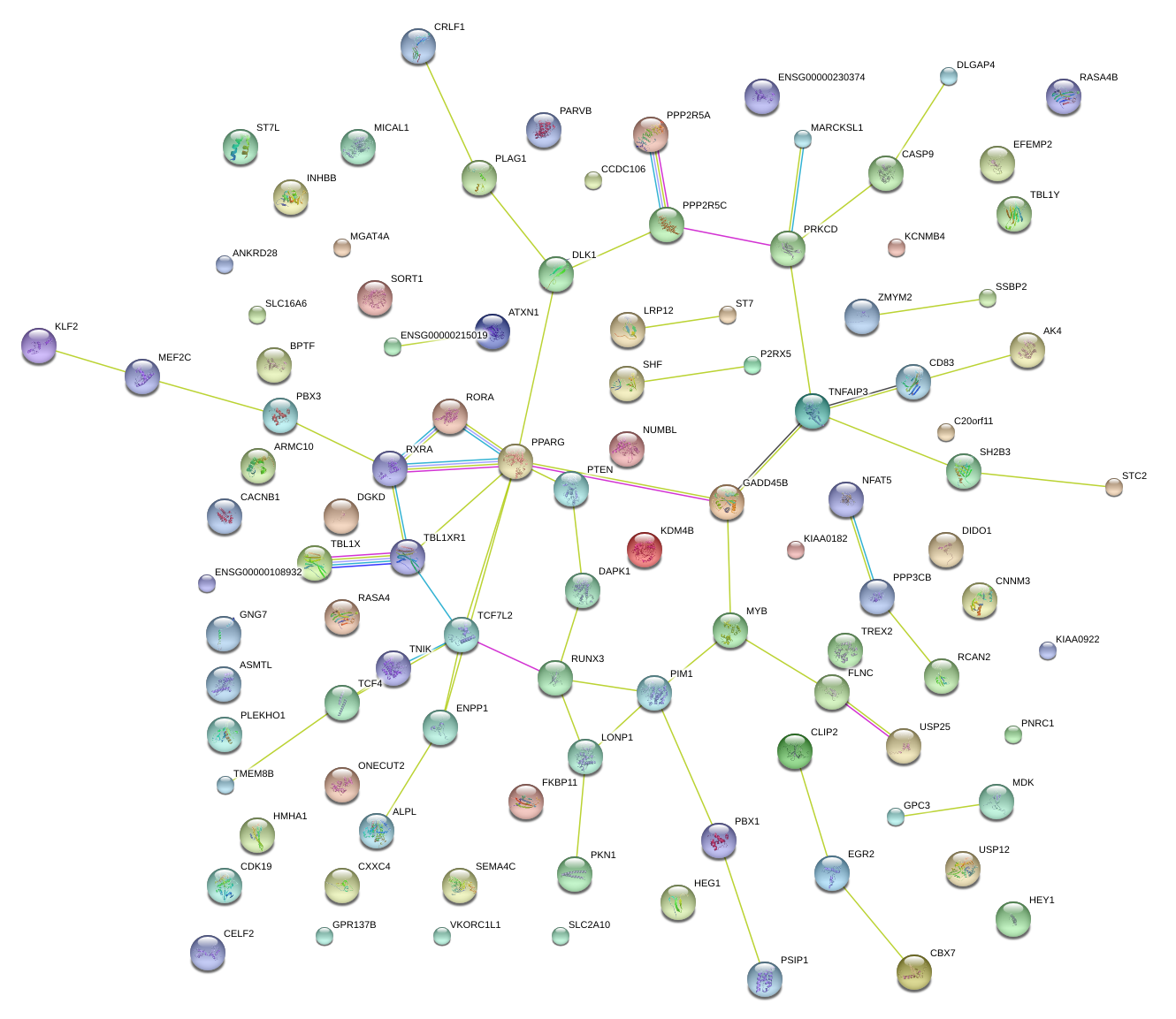
|
chr1_-_32574185
|
1.054
|
NM_023009
|
MARCKSL1
|
MARCKS-like 1
|
|
chr19_+_16296632
|
0.668
|
NM_016270
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr19_+_16296734
|
0.648
|
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr2_+_233927892
|
0.580
|
|
DGKD
|
diacylglycerol kinase, delta 130kDa
|
|
chr17_-_76622967
|
0.555
|
|
FLJ90757
|
hypothetical LOC440465
|
|
chr9_+_127549437
|
0.526
|
NM_006195
|
PBX3
|
pre-B-cell leukemia homeobox 3
|
|
chr3_-_172660545
|
0.509
|
NM_001161560
NM_001161561
NM_001161562
NM_001161563
NM_001161564
NM_001161565
NM_001161566
NM_015028
|
TNIK
|
TRAF2 and NCK interacting kinase
|
|
chr10_+_11099860
|
0.492
|
NM_006561
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr5_-_88215034
|
0.468
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr19_+_4920126
|
0.463
|
|
KDM4B
|
lysine (K)-specific demethylase 4B
|
|
chr10_+_11100075
|
0.456
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr1_+_148388870
|
0.448
|
|
PLEKHO1
|
pleckstrin homology domain containing, family O member 1
|
|
chr1_-_25128951
|
0.446
|
|
RUNX3
|
runt-related transcription factor 3
|
|
chr10_-_74925724
|
0.440
|
|
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme
|
|
chr16_+_84204424
|
0.423
|
NM_014615
|
KIAA0182
|
KIAA0182
|
|
chr1_+_148389039
|
0.411
|
|
PLEKHO1
|
pleckstrin homology domain containing, family O member 1
|
|
chr15_-_58671929
|
0.401
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr19_-_2653738
|
0.398
|
NM_052847
|
GNG7
|
guanine nucleotide binding protein (G protein), gamma 7
|
|
chr12_+_69046241
|
0.392
|
NM_014505
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4
|
|
chr16_+_29725597
|
0.382
|
|
|
|
|
chrY_-_1531733
|
0.369
|
NM_001173474
NM_004192
|
ASMTL
|
acetylserotonin O-methyltransferase-like
|
|
chrX_-_1531733
|
0.369
|
NM_001173474
NM_004192
|
ASMTL
|
acetylserotonin O-methyltransferase-like
|
|
chr9_-_15500899
|
0.366
|
NM_001128217
NM_033222
|
PSIP1
|
PC4 and SFRS1 interacting protein 1
|
|
chrX_+_9392980
|
0.363
|
NM_005647
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr10_+_89613399
|
0.362
|
|
PTEN
|
phosphatase and tensin homolog
|
|
chr2_+_233927880
|
0.361
|
NM_152879
|
DGKD
|
diacylglycerol kinase, delta 130kDa
|
|
chr16_+_29725665
|
0.354
|
|
|
|
|
chr8_-_57286381
|
0.353
|
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chr15_-_59308708
|
0.346
|
|
RORA
|
RAR-related orphan receptor A
|
|
chr10_-_64248932
|
0.344
|
NM_001136178
|
EGR2
|
early growth response 2
|
|
chr10_+_89613071
|
0.335
|
NM_000314
|
PTEN
|
phosphatase and tensin homolog
|
|
chr17_-_63798851
|
0.334
|
NM_001174166
NM_004694
|
SLC16A6
|
solute carrier family 16, member 6 (monocarboxylic acid transporter 7)
|
|
chr20_+_34357604
|
0.333
|
|
DLGAP4
|
discs, large (Drosophila) homolog-associated protein 4
|
|
chr11_+_46359878
|
0.331
|
NM_001012333
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr15_-_59308770
|
0.330
|
NM_134261
|
RORA
|
RAR-related orphan receptor A
|
|
chr17_+_63252668
|
0.327
|
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr19_+_1022208
|
0.325
|
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr19_-_18578467
|
0.325
|
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr6_+_37245860
|
0.318
|
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr9_+_136358145
|
0.317
|
|
RXRA
|
retinoid X receptor, alpha
|
|
chrX_-_132947290
|
0.313
|
NM_001164617
NM_001164618
NM_001164619
NM_004484
|
GPC3
|
glypican 3
|
|
chr4_+_154606900
|
0.312
|
NM_001131007
NM_015196
|
KIAA0922
|
KIAA0922
|
|
chr4_-_105631915
|
0.312
|
NM_025212
|
CXXC4
|
CXXC finger protein 4
|
|
chr6_-_46566969
|
0.308
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chr7_+_64975596
|
0.308
|
NM_173517
|
VKORC1L1
|
vitamin K epoxide reductase complex, subunit 1-like 1
|
|
chr22_-_37878394
|
0.307
|
NM_175709
|
CBX7
|
chromobox homolog 7
|
|
chr1_-_15723363
|
0.302
|
NM_001229
NM_032996
|
CASP9
|
caspase 9, apoptosis-related cysteine peptidase
|
|
chr20_+_61039803
|
0.302
|
NM_017896
|
C20orf11
|
chromosome 20 open reading frame 11
|
|
chr9_+_35819208
|
0.299
|
NM_016446
NM_001042589
|
TMEM8B
|
transmembrane protein 8B
|
|
chr13_+_19430788
|
0.298
|
NM_003453
NM_197968
NM_001190964
|
ZMYM2
|
zinc finger, MYM-type 2
|
|
chr10_+_11100111
|
0.296
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr6_+_132170888
|
0.296
|
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chr11_+_46359821
|
0.296
|
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr7_+_102502852
|
0.294
|
|
ARMC10
|
armadillo repeat containing 10
|
|
chr20_-_61039672
|
0.293
|
|
DIDO1
|
death inducer-obliterator 1
|
|
chr16_+_68157611
|
0.292
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chrX_-_132947141
|
0.290
|
|
GPC3
|
glypican 3
|
|
chr11_+_46359781
|
0.290
|
NM_002391
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr9_+_35819375
|
0.289
|
NM_001042590
|
TMEM8B
|
transmembrane protein 8B
|
|
chr19_-_2653662
|
0.289
|
|
GNG7
|
guanine nucleotide binding protein (G protein), gamma 7
|
|
chr17_-_3546047
|
0.288
|
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5
|
|
chr6_+_138230045
|
0.281
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr11_+_46359855
|
0.276
|
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr19_+_2427122
|
0.275
|
NM_015675
|
GADD45B
|
growth arrest and DNA-damage-inducible, beta
|
|
chr6_-_109883864
|
0.275
|
NM_001159291
NM_022765
|
MICAL1
|
microtubule associated monoxygenase, calponin and LIM domain containing 1
|
|
chr6_-_109883635
|
0.274
|
|
MICAL1
|
microtubule associated monoxygenase, calponin and LIM domain containing 1
|
|
chr16_+_65018316
|
0.270
|
NM_001136106
NM_001178020
NM_001197224
NM_001197225
|
BEAN1
|
brain expressed, associated with NEDD4, 1
|
|
chr13_+_19430906
|
0.269
|
NM_001190965
|
ZMYM2
|
zinc finger, MYM-type 2
|
|
chr11_+_46359836
|
0.269
|
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr5_-_172688024
|
0.268
|
|
STC2
|
stanniocalcin 2
|
|
chr6_+_135544170
|
0.268
|
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr6_-_111243023
|
0.264
|
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr22_+_42751487
|
0.264
|
NM_013327
|
PARVB
|
parvin, beta
|
|
chr5_-_81082699
|
0.263
|
NM_012446
|
SSBP2
|
single-stranded DNA binding protein 2
|
|
chr6_+_89848229
|
0.263
|
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr9_+_89302239
|
0.261
|
|
DAPK1
|
death-associated protein kinase 1
|
|
chr19_-_18578631
|
0.261
|
NM_004750
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr2_+_96845704
|
0.261
|
NM_017623
NM_199078
|
CNNM3
|
cyclin M3
|
|
chr6_-_109883778
|
0.261
|
|
MICAL1
|
microtubule associated monoxygenase, calponin and LIM domain containing 1
|
|
chr18_-_51406359
|
0.261
|
|
TCF4
|
transcription factor 4
|
|
chr17_-_3546236
|
0.260
|
NM_002561
NM_175080
NM_175081
|
P2RX5-TAX1BP3
P2RX5
|
P2RX5-TAX1BP3 read-through transcript
purinergic receptor P2X, ligand-gated ion channel, 5
|
|
chr13_-_26643952
|
0.258
|
NM_182488
|
USP12
|
ubiquitin specific peptidase 12
|
|
chr6_-_16869579
|
0.258
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chr19_+_60851207
|
0.257
|
|
CCDC106
|
coiled-coil domain containing 106
|
|
chr6_+_135544138
|
0.257
|
NM_001130172
NM_001130173
NM_001161656
NM_001161657
NM_001161658
NM_001161659
NM_001161660
NM_005375
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr7_+_128257698
|
0.256
|
NM_001127487
NM_001458
|
FLNC
|
filamin C, gamma
|
|
chr7_-_102044419
|
0.255
|
NM_001079877
NM_006989
|
RASA4
|
RAS p21 protein activator 4
|
|
chr7_-_102044374
|
0.255
|
|
RASA4
|
RAS p21 protein activator 4
|
|
chr1_+_148388719
|
0.255
|
NM_016274
|
PLEKHO1
|
pleckstrin homology domain containing, family O member 1
|
|
chr15_-_43267441
|
0.254
|
|
SHF
|
Src homology 2 domain containing F
|
|
chr19_+_14405216
|
0.253
|
|
PKN1
|
protein kinase N1
|
|
chr3_-_15875932
|
0.252
|
NM_015199
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr13_+_19430881
|
0.251
|
|
ZMYM2
|
zinc finger, MYM-type 2
|
|
chr6_+_89847183
|
0.250
|
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr2_-_96899396
|
0.250
|
NM_017789
|
SEMA4C
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C
|
|
chr14_+_101297887
|
0.248
|
NM_001161725
NM_001161726
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr1_+_65385799
|
0.247
|
NM_203464
NM_001005353
|
AK4
|
adenylate kinase 4
|
|
chr10_+_89613347
|
0.247
|
|
PTEN
|
phosphatase and tensin homolog
|
|
chr14_+_100262916
|
0.246
|
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chr6_-_109883639
|
0.246
|
|
MICAL1
|
microtubule associated monoxygenase, calponin and LIM domain containing 1
|
|
chr1_+_234372454
|
0.245
|
NM_003272
|
GPR137B
|
G protein-coupled receptor 137B
|
|
chr7_+_73341640
|
0.245
|
NM_003388
NM_032421
|
CLIP2
|
CAP-GLY domain containing linker protein 2
|
|
chr8_-_57286412
|
0.244
|
NM_001114634
NM_001114635
NM_002655
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chr20_-_61039710
|
0.242
|
NM_022105
NM_033081
NM_080797
|
DIDO1
|
death inducer-obliterator 1
|
|
chr18_+_53253775
|
0.242
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|
|
chr7_+_116380790
|
0.241
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr19_-_45888259
|
0.240
|
NM_004756
|
NUMBL
|
numb homolog (Drosophila)-like
|
|
chr21_+_16024241
|
0.236
|
NM_013396
|
USP25
|
ubiquitin specific peptidase 25
|
|
chr11_-_65396775
|
0.235
|
NM_016938
|
EFEMP2
|
EGF containing fibulin-like extracellular matrix protein 2
|
|
chr1_+_210525885
|
0.234
|
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha
|
|
chr14_+_100263066
|
0.232
|
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chr3_+_12304332
|
0.232
|
NM_005037
NM_138712
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr9_-_15500209
|
0.231
|
NM_021144
|
PSIP1
|
PC4 and SFRS1 interacting protein 1
|
|
chr1_+_210525792
|
0.231
|
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha
|
|
chr7_+_102502816
|
0.230
|
|
ARMC10
|
armadillo repeat containing 10
|
|
chr3_-_126257425
|
0.230
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr2_-_98713862
|
0.229
|
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr17_+_63252704
|
0.229
|
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr1_-_109742020
|
0.228
|
NM_002959
|
SORT1
|
sortilin 1
|
|
chr20_+_44771532
|
0.228
|
NM_030777
|
SLC2A10
|
solute carrier family 2 (facilitated glucose transporter), member 10
|
|
chr12_+_110328126
|
0.228
|
NM_005475
|
SH2B3
|
SH2B adaptor protein 3
|
|
chr2_+_120820140
|
0.227
|
NM_002193
|
INHBB
|
inhibin, beta B
|
|
chr6_+_14225836
|
0.227
|
NM_001040280
NM_004233
|
CD83
|
CD83 molecule
|
|
chr13_-_26643775
|
0.226
|
|
USP12
|
ubiquitin specific peptidase 12
|
|
chr1_+_21708502
|
0.223
|
|
ALPL
|
alkaline phosphatase, liver/bone/kidney
|
|
chr4_-_74343169
|
0.222
|
|
|
|
|
chr3_+_53170254
|
0.221
|
NM_006254
NM_212539
|
PRKCD
|
protein kinase C, delta
|
|
chr8_-_80842452
|
0.220
|
NM_001040708
NM_012258
|
HEY1
|
hairy/enhancer-of-split related with YRPW motif 1
|
|
chr2_-_225615556
|
0.220
|
NM_014689
|
DOCK10
|
dedicator of cytokinesis 10
|
|
chr10_+_69314347
|
0.218
|
NM_012238
|
SIRT1
|
sirtuin 1
|
|
chrX_+_46977306
|
0.217
|
|
USP11
|
ubiquitin specific peptidase 11
|
|
chr2_+_203901160
|
0.217
|
NM_005759
|
ABI2
|
abl-interactor 2
|
|
chr16_+_1143241
|
0.217
|
NM_001005407
NM_021098
|
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit
|
|
chr11_-_92916161
|
0.216
|
NM_020179
|
C11orf75
|
chromosome 11 open reading frame 75
|
|
chr4_+_25931725
|
0.215
|
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region
|
|
chr7_+_119700924
|
0.214
|
NM_012281
|
KCND2
|
potassium voltage-gated channel, Shal-related subfamily, member 2
|
|
chr10_+_69314714
|
0.214
|
|
SIRT1
|
sirtuin 1
|
|
chr19_+_18069596
|
0.213
|
NM_015016
|
MAST3
|
microtubule associated serine/threonine kinase 3
|
|
chr10_+_45189673
|
0.212
|
|
ALOX5
|
arachidonate 5-lipoxygenase
|
|
chr8_-_72918938
|
0.212
|
|
MSC
|
musculin
|
|
chr4_+_25931534
|
0.211
|
NM_015874
NM_203284
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region
|
|
chr6_+_132170847
|
0.208
|
NM_006208
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chr6_-_111243100
|
0.207
|
NM_015076
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr19_-_18253436
|
0.207
|
|
JUND
|
jun D proto-oncogene
|
|
chr10_+_45189617
|
0.207
|
NM_000698
|
ALOX5
|
arachidonate 5-lipoxygenase
|
|
chr14_+_99220349
|
0.204
|
NM_006668
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1
|
|
chr6_-_111911106
|
0.203
|
NM_002912
|
REV3L
|
REV3-like, catalytic subunit of DNA polymerase zeta (yeast)
|
|
chr17_-_8006979
|
0.203
|
NM_014232
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2)
|
|
chr19_+_659766
|
0.201
|
NM_001040134
NM_002579
|
PALM
|
paralemmin
|
|
chr9_-_36390817
|
0.200
|
NM_194328
NM_194332
|
RNF38
|
ring finger protein 38
|
|
chr6_-_112301123
|
0.196
|
NM_002037
|
FYN
|
FYN oncogene related to SRC, FGR, YES
|
|
chr8_-_74821610
|
0.196
|
|
STAU2
LOC729696
|
staufen, RNA binding protein, homolog 2 (Drosophila)
hypothetical protein LOC729696
|
|
chr5_-_81082614
|
0.196
|
|
SSBP2
|
single-stranded DNA binding protein 2
|
|
chr19_+_4920123
|
0.195
|
NM_015015
|
KDM4B
|
lysine (K)-specific demethylase 4B
|
|
chr1_-_149698272
|
0.195
|
|
POGZ
|
pogo transposable element with ZNF domain
|
|
chr6_+_71434305
|
0.194
|
|
SMAP1
|
small ArfGAP 1
|
|
chr19_+_10843244
|
0.194
|
NM_199141
|
CARM1
|
coactivator-associated arginine methyltransferase 1
|
|
chr16_-_3870686
|
0.194
|
|
CREBBP
|
CREB binding protein
|
|
chr8_-_72919161
|
0.193
|
|
MSC
|
musculin
|
|
chr6_-_143307976
|
0.193
|
NM_006734
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chr17_-_74432671
|
0.192
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr18_-_24011349
|
0.192
|
NM_001792
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr13_-_40138607
|
0.191
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr2_+_86521781
|
0.191
|
NM_018433
|
KDM3A
|
lysine (K)-specific demethylase 3A
|
|
chr22_+_38183262
|
0.189
|
NM_002409
|
MGAT3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase
|
|
chr1_+_209566789
|
0.188
|
|
TRAF5
|
TNF receptor-associated factor 5
|
|
chr17_-_40750109
|
0.188
|
NM_003954
|
MAP3K14
|
mitogen-activated protein kinase kinase kinase 14
|
|
chr1_+_209566770
|
0.188
|
NM_001033910
|
TRAF5
|
TNF receptor-associated factor 5
|
|
chr1_-_149698516
|
0.187
|
NM_001194937
NM_001194938
NM_015100
NM_145796
|
POGZ
|
pogo transposable element with ZNF domain
|
|
chr19_-_44032388
|
0.186
|
|
HNRNPL
|
heterogeneous nuclear ribonucleoprotein L
|
|
chr4_+_37569093
|
0.186
|
NM_015173
|
TBC1D1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1
|
|
chr6_+_42857540
|
0.185
|
|
KIAA0240
|
KIAA0240
|
|
chr17_+_63252087
|
0.184
|
NM_004459
NM_182641
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr11_+_66790517
|
0.184
|
|
ADRBK1
|
adrenergic, beta, receptor kinase 1
|
|
chr2_+_162908869
|
0.182
|
|
GCA
|
grancalcin, EF-hand calcium binding protein
|
|
chr17_-_15407497
|
0.181
|
NM_001135036
NM_145301
|
FAM18B2-CDRT4
FAM18B2
|
FAM18B2-CDRT4 read-through transcript
family with sequence similarity 18, member B2
|
|
chr7_+_102502529
|
0.180
|
NM_001161009
NM_001161010
NM_001161011
NM_001161012
NM_001161013
NM_031905
|
ARMC10
|
armadillo repeat containing 10
|
|
chr17_-_78602931
|
0.179
|
NM_001009905
|
B3GNTL1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1
|
|
chr20_-_61600834
|
0.178
|
NM_001958
|
EEF1A2
|
eukaryotic translation elongation factor 1 alpha 2
|
|
chr1_-_198905619
|
0.177
|
|
DDX59
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 59
|
|
chr12_+_102982365
|
0.177
|
NM_013320
|
HCFC2
|
host cell factor C2
|
|
chr3_-_38666122
|
0.175
|
NM_000335
NM_001099404
NM_001099405
NM_198056
NM_001160160
NM_001160161
|
SCN5A
|
sodium channel, voltage-gated, type V, alpha subunit
|
|
chr8_+_38877909
|
0.175
|
NM_021623
|
PLEKHA2
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2
|
|
chr2_+_54536780
|
0.175
|
NM_003128
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|
|
chr5_-_39110235
|
0.173
|
|
RICTOR
|
RPTOR independent companion of MTOR, complex 2
|
|
chr1_+_110012166
|
0.173
|
NM_000848
NM_001142368
|
GSTM2
|
glutathione S-transferase mu 2 (muscle)
|
|
chr14_+_105024301
|
0.173
|
NM_001311
|
CRIP1
|
cysteine-rich protein 1 (intestinal)
|
|
chr14_+_105024576
|
0.172
|
|
CRIP1
|
cysteine-rich protein 1 (intestinal)
|
|
chr5_-_172687798
|
0.172
|
|
STC2
|
stanniocalcin 2
|
|
chr9_+_2005218
|
0.172
|
NM_003070
NM_139045
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2
|
|
chr8_-_74821636
|
0.172
|
NM_001164380
NM_001164381
NM_001164382
NM_001164383
NM_001164385
NM_014393
|
STAU2
|
staufen, RNA binding protein, homolog 2 (Drosophila)
|
|
chr17_-_74432800
|
0.172
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr7_+_127883117
|
0.172
|
NM_001098786
NM_013332
|
C7orf68
|
chromosome 7 open reading frame 68
|
|
chr7_+_149696901
|
0.170
|
|
REPIN1
|
replication initiator 1
|
|
chr12_-_99060590
|
0.170
|
|
UHRF1BP1L
|
UHRF1 binding protein 1-like
|
|
chr20_+_60130866
|
0.168
|
NM_144703
|
LSM14B
|
LSM14B, SCD6 homolog B (S. cerevisiae)
|
|
chr7_-_130891861
|
0.168
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr16_-_960982
|
0.168
|
NM_022773
|
LMF1
|
lipase maturation factor 1
|
|
chr6_-_79844606
|
0.168
|
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr11_-_9243408
|
0.167
|
NM_015213
|
DENND5A
|
DENN/MADD domain containing 5A
|