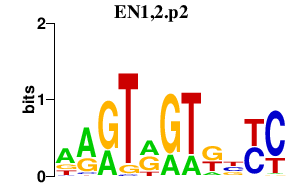
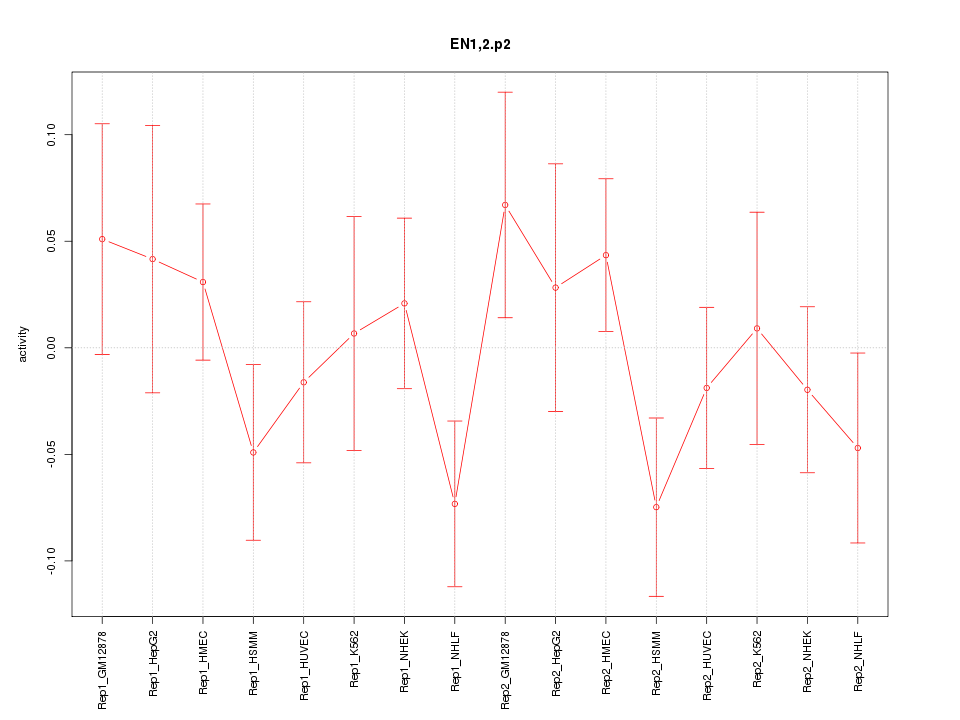
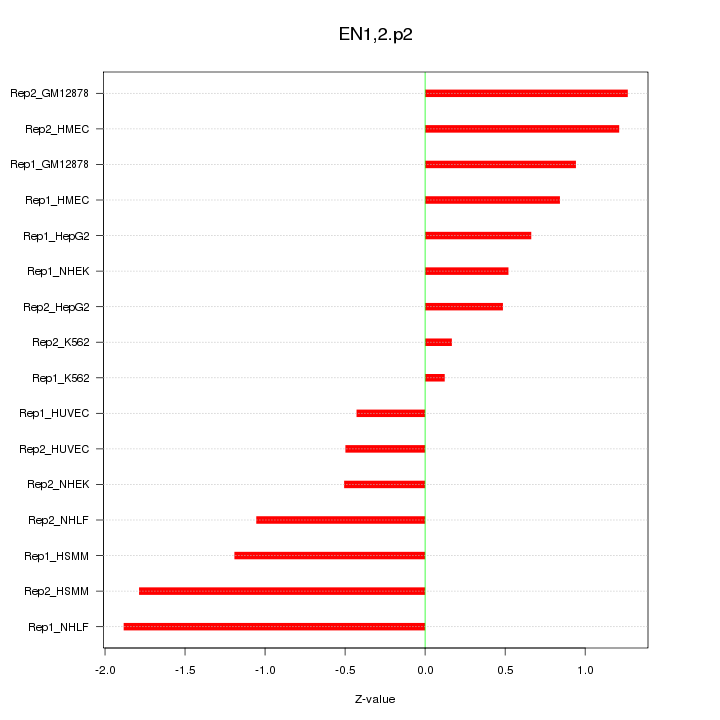
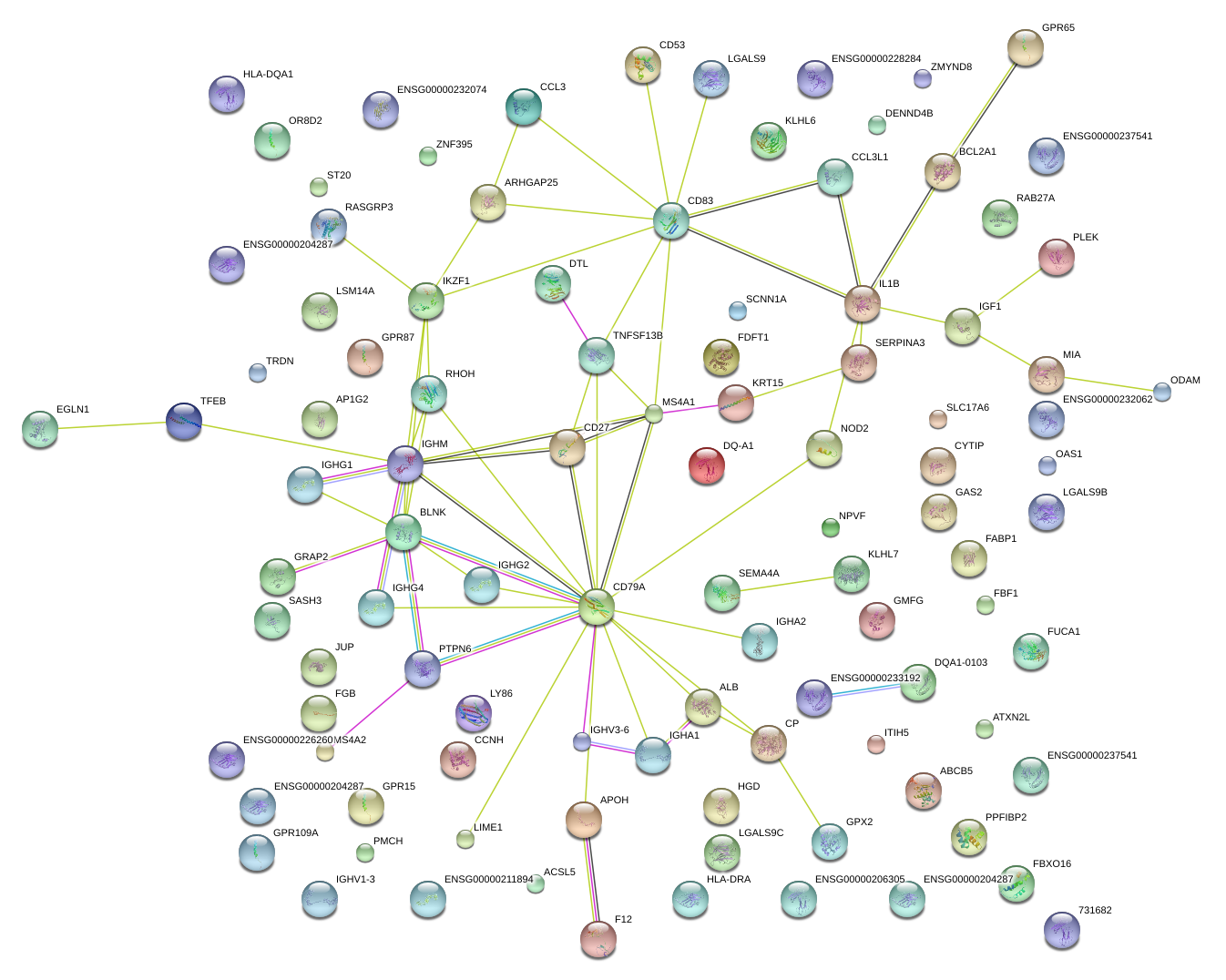
|
chr10_-_98021144
|
2.567
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr4_+_74500748
|
2.466
|
|
ALB
|
albumin
|
|
chr11_+_22652894
|
2.424
|
NM_005256
|
GAS2
|
growth arrest-specific 2
|
|
chr11_+_59979857
|
1.516
|
NM_021950
NM_152866
|
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1
|
|
chr4_+_74488947
|
1.438
|
|
ALB
|
albumin
|
|
chr17_-_36928604
|
1.415
|
NM_002275
|
KRT15
|
keratin 15
|
|
chr14_-_105182025
|
1.169
|
|
|
|
|
chr6_+_32515596
|
1.106
|
NM_019111
|
HLA-DRA
|
major histocompatibility complex, class II, DR alpha
|
|
chr2_-_158008763
|
1.081
|
NM_004288
|
CYTIP
|
cytohesin 1 interacting protein
|
|
chr17_-_61655915
|
0.936
|
NM_000042
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I)
|
|
chr14_-_105845570
|
0.930
|
|
IGHA1
|
immunoglobulin heavy constant alpha 1
|
|
chr2_-_88208650
|
0.913
|
NM_001443
|
FABP1
|
fatty acid binding protein 1, liver
|
|
chr10_+_114123828
|
0.903
|
NM_203379
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr15_-_78050515
|
0.877
|
NM_001114735
NM_004049
|
BCL2A1
|
BCL2-related protein A1
|
|
chr12_-_101115677
|
0.869
|
NM_002674
|
PMCH
|
pro-melanin-concentrating hormone
|
|
chr12_+_111829191
|
0.848
|
|
OAS1
|
2',5'-oligoadenylate synthetase 1, 40/46kDa
|
|
chr17_-_31441390
|
0.836
|
NM_002983
|
CCL3
|
chemokine (C-C motif) ligand 3
|
|
chr17_-_37181636
|
0.821
|
|
JUP
|
junction plakoglobin
|
|
chr12_+_6424293
|
0.818
|
NM_001242
|
CD27
|
CD27 molecule
|
|
chr14_-_64479198
|
0.784
|
NM_002083
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal)
|
|
chr6_-_41811916
|
0.766
|
NM_001167827
|
TFEB
|
transcription factor EB
|
|
chr1_+_111217244
|
0.727
|
NM_000560
|
CD53
|
CD53 molecule
|
|
chr12_-_6356783
|
0.696
|
NM_001159575
|
SCNN1A
|
sodium channel, nonvoltage-gated 1 alpha
|
|
chr14_-_105182159
|
0.690
|
|
IGHG2
|
immunoglobulin heavy constant gamma 2 (G2m marker)
|
|
chr17_+_22996461
|
0.679
|
|
LGALS9
|
lectin, galactoside-binding, soluble, 9
|
|
chr3_-_184756101
|
0.670
|
NM_130446
|
KLHL6
|
kelch-like 6 (Drosophila)
|
|
chr4_+_39874921
|
0.658
|
NM_004310
|
RHOH
|
ras homolog gene family, member H
|
|
chr2_+_33592445
|
0.650
|
NM_015376
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr5_-_176769141
|
0.633
|
NM_000505
|
F12
|
coagulation factor XII (Hageman factor)
|
|
chr2_+_68855436
|
0.620
|
NM_001166276
NM_014882
|
ARHGAP25
|
Rho GTPase activating protein 25
|
|
chr6_+_30402203
|
0.608
|
|
|
|
|
chr14_+_87541220
|
0.603
|
NM_003608
|
GPR65
|
G protein-coupled receptor 65
|
|
chr22_+_38627031
|
0.602
|
NM_004810
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr2_-_113310796
|
0.583
|
NM_000576
|
IL1B
|
interleukin 1, beta
|
|
chr20_-_45410033
|
0.583
|
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr3_-_150422474
|
0.578
|
NM_000096
|
CP
|
ceruloplasmin (ferroxidase)
|
|
chr7_+_20653490
|
0.551
|
NM_001163942
NM_001163993
NM_178559
|
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5
|
|
chr6_+_32713160
|
0.548
|
NM_002122
|
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1
|
|
chr8_-_28403652
|
0.530
|
NM_172366
|
FBXO16
ZNF395
|
F-box protein 16
zinc finger protein 395
|
|
chr10_+_114123765
|
0.527
|
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr3_-_121884091
|
0.523
|
NM_000187
|
HGD
|
homogentisate 1,2-dioxygenase
|
|
chr11_+_7574992
|
0.510
|
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr4_+_71096830
|
0.499
|
NM_017855
|
ODAM
|
odontogenic, ameloblast asssociated
|
|
chr15_-_53350383
|
0.486
|
NM_183236
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr19_+_45973139
|
0.481
|
NM_006533
|
MIA
|
melanoma inhibitory activity
|
|
chr6_+_14225836
|
0.480
|
NM_001040280
NM_004233
|
CD83
|
CD83 molecule
|
|
chr8_-_38577891
|
0.479
|
|
|
|
|
chr5_-_86739683
|
0.468
|
|
CCNH
|
cyclin H
|
|
chrX_+_128741578
|
0.466
|
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr12_+_111829121
|
0.463
|
NM_001032409
NM_002534
NM_016816
|
OAS1
|
2',5'-oligoadenylate synthetase 1, 40/46kDa
|
|
chr20_+_61838479
|
0.461
|
|
LIME1
|
Lck interacting transmembrane adaptor 1
|
|
chr2_+_68445936
|
0.456
|
|
PLEK
|
pleckstrin
|
|
chr14_-_23107106
|
0.454
|
NM_003917
|
AP1G2
|
adaptor-related protein complex 1, gamma 2 subunit
|
|
chr4_+_155703581
|
0.450
|
NM_001184741
NM_005141
|
FGB
|
fibrinogen beta chain
|
|
chr19_-_44518465
|
0.450
|
|
GMFG
|
glia maturation factor, gamma
|
|
chr6_+_6533932
|
0.449
|
NM_004271
|
LY86
|
lymphocyte antigen 86
|
|
chr4_+_74488835
|
0.446
|
NM_000477
|
ALB
|
albumin
|
|
chr3_-_152517204
|
0.441
|
NM_023915
|
GPR87
|
G protein-coupled receptor 87
|
|
chrX_+_128741562
|
0.439
|
NM_018990
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr22_+_38627129
|
0.433
|
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr19_-_44518516
|
0.421
|
NM_004877
|
GMFG
|
glia maturation factor, gamma
|
|
chr6_-_123999640
|
0.419
|
NM_006073
|
TRDN
|
triadin
|
|
chr14_-_106149623
|
0.406
|
|
|
|
|
chr1_+_210275752
|
0.405
|
|
DTL
|
denticleless homolog (Drosophila)
|
|
chr1_+_154390939
|
0.402
|
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr20_+_61838389
|
0.399
|
NM_017806
|
LIME1
|
Lck interacting transmembrane adaptor 1
|
|
chr12_-_121767272
|
0.395
|
NM_006018
|
GPR109B
|
G protein-coupled receptor 109B
|
|
chr19_+_47073029
|
0.384
|
NM_001783
NM_021601
|
CD79A
|
CD79a molecule, immunoglobulin-associated alpha
|
|
chr11_+_22316242
|
0.384
|
NM_020346
|
SLC17A6
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6
|
|
chr1_-_24067347
|
0.383
|
NM_000147
|
FUCA1
|
fucosidase, alpha-L- 1, tissue
|
|
chr8_+_11702803
|
0.381
|
|
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1
|
|
chr12_+_6925997
|
0.378
|
NM_080548
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr14_+_94148466
|
0.376
|
NM_001085
|
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3
|
|
chr1_-_229627412
|
0.374
|
NM_022051
|
EGLN1
|
egl nine homolog 1 (C. elegans)
|
|
chr3_+_99733432
|
0.374
|
NM_005290
|
GPR15
|
G protein-coupled receptor 15
|
|
chr7_-_25234582
|
0.373
|
NM_022150
|
NPVF
|
neuropeptide VF precursor
|
|
chr22_+_21053972
|
0.369
|
|
|
|
|
chr16_+_49288550
|
0.364
|
NM_022162
|
NOD2
|
nucleotide-binding oligomerization domain containing 2
|
|
chr12_-_101396459
|
0.362
|
NM_001111284
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr12_+_6926014
|
0.361
|
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr7_+_50318801
|
0.356
|
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr18_-_28247186
|
0.354
|
|
|
|
|
chr19_+_39402167
|
0.351
|
|
LSM14A
|
LSM14A, SCD6 homolog A (S. cerevisiae)
|
|
chr1_-_152185764
|
0.350
|
NM_014856
|
DENND4B
|
DENN/MADD domain containing 4B
|
|
chr10_-_7722747
|
0.349
|
|
ITIH5
|
inter-alpha (globulin) inhibitor H5
|
|
chr14_-_106038573
|
0.347
|
|
IGHA1
IGHA2
IGHG1
IGHM
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant mu
|
|
chr16_+_28850760
|
0.342
|
NM_001178098
NM_001770
|
CD19
|
CD19 molecule
|
|
chr2_+_102401580
|
0.342
|
NM_003853
|
IL18RAP
|
interleukin 18 receptor accessory protein
|
|
chr1_+_31823097
|
0.341
|
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr17_-_36347197
|
0.341
|
NM_015515
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr11_+_22646229
|
0.339
|
NM_177553
|
GAS2
|
growth arrest-specific 2
|
|
chr1_-_108032645
|
0.333
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr6_-_79732391
|
0.332
|
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr19_-_56164628
|
0.331
|
NM_002774
|
KLK6
|
kallikrein-related peptidase 6
|
|
chr17_-_53946899
|
0.331
|
|
MTMR4
|
myotubularin related protein 4
|
|
chr2_+_171316462
|
0.329
|
|
EIF2S2
|
eukaryotic translation initiation factor 2, subunit 2 beta, 38kDa
|
|
chr4_-_77147625
|
0.324
|
NM_002416
|
CXCL9
|
chemokine (C-X-C motif) ligand 9
|
|
chr10_-_105835525
|
0.323
|
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr6_+_106653429
|
0.321
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr4_+_74493266
|
0.320
|
|
ALB
|
albumin
|
|
chr15_-_50375143
|
0.317
|
NM_018728
|
MYO5C
|
myosin VC
|
|
chr14_-_93924873
|
0.316
|
NM_000295
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr1_-_23998515
|
0.313
|
NM_001127621
|
GALE
|
UDP-galactose-4-epimerase
|
|
chrX_-_48655978
|
0.308
|
|
PIM2
|
pim-2 oncogene
|
|
chr16_+_55950195
|
0.308
|
NM_002990
|
CCL22
|
chemokine (C-C motif) ligand 22
|
|
chr7_+_106897737
|
0.305
|
NM_005295
|
GPR22
|
G protein-coupled receptor 22
|
|
chr6_+_32021831
|
0.304
|
|
CFB
|
complement factor B
|
|
chr5_-_176769094
|
0.304
|
|
F12
|
coagulation factor XII (Hageman factor)
|
|
chr3_-_122862391
|
0.303
|
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr12_-_101036407
|
0.303
|
NM_024057
|
NUP37
|
nucleoporin 37kDa
|
|
chr4_-_17415918
|
0.302
|
|
DCAF16
|
DDB1 and CUL4 associated factor 16
|
|
chr12_+_84792203
|
0.299
|
NM_006183
|
NTS
|
neurotensin
|
|
chr1_-_159459999
|
0.294
|
NM_001643
|
APOA2
|
apolipoprotein A-II
|
|
chr17_-_24491521
|
0.293
|
|
MYO18A
|
myosin XVIIIA
|
|
chr15_+_41672543
|
0.292
|
NM_020990
|
CKMT1B
CKMT1A
|
creatine kinase, mitochondrial 1B
creatine kinase, mitochondrial 1A
|
|
chr1_+_20788027
|
0.288
|
NM_001785
|
CDA
|
cytidine deaminase
|
|
chr5_-_169749215
|
0.288
|
NM_004137
|
KCNMB1
|
potassium large conductance calcium-activated channel, subfamily M, beta member 1
|
|
chr14_-_94669547
|
0.288
|
NM_001195573
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr9_-_103185621
|
0.287
|
NM_001127610
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase)
|
|
chr4_+_74499651
|
0.282
|
|
ALB
|
albumin
|
|
chr1_+_210275541
|
0.275
|
NM_016448
|
DTL
|
denticleless homolog (Drosophila)
|
|
chr3_-_122862447
|
0.275
|
NM_005335
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr6_-_133126278
|
0.275
|
NM_078488
|
VNN2
|
vanin 2
|
|
chr1_+_111571803
|
0.272
|
NM_001025197
NM_004000
|
CHI3L2
|
chitinase 3-like 2
|
|
chr7_+_115650240
|
0.272
|
NM_152829
|
TES
|
testis derived transcript (3 LIM domains)
|
|
chr5_+_68703036
|
0.269
|
NM_133339
|
RAD17
|
RAD17 homolog (S. pombe)
|
|
chr13_-_45654296
|
0.269
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr12_-_51585004
|
0.267
|
NM_002273
|
KRT8
|
keratin 8
|
|
chr7_+_94130752
|
0.266
|
|
PEG10
|
paternally expressed 10
|
|
chr13_-_45577144
|
0.266
|
NM_001872
NM_016413
|
CPB2
|
carboxypeptidase B2 (plasma)
|
|
chr3_-_121883647
|
0.261
|
|
HGD
|
homogentisate 1,2-dioxygenase
|
|
chr12_-_61615124
|
0.260
|
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr17_-_26665144
|
0.260
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr14_+_59782222
|
0.257
|
NM_177952
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A
|
|
chr12_+_68028407
|
0.256
|
|
LYZ
|
lysozyme
|
|
chr3_+_42176662
|
0.255
|
NM_014965
|
TRAK1
|
trafficking protein, kinesin binding 1
|
|
chr9_-_122852356
|
0.253
|
NM_001735
|
C5
|
complement component 5
|
|
chr6_-_154692906
|
0.251
|
NM_001130699
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1
|
|
chr1_+_86662356
|
0.250
|
NM_006536
|
CLCA2
|
chloride channel accessory 2
|
|
chrX_-_47374647
|
0.250
|
NM_002621
|
CFP
|
complement factor properdin
|
|
chr14_-_105280425
|
0.248
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr16_+_2593351
|
0.248
|
|
LOC652276
|
hypothetical LOC652276
|
|
chr6_+_12825818
|
0.248
|
NM_030948
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr18_+_27425727
|
0.247
|
NM_000371
|
TTR
|
transthyretin
|
|
chr9_-_122728993
|
0.246
|
NM_005658
|
TRAF1
|
TNF receptor-associated factor 1
|
|
chr15_-_32416336
|
0.245
|
NM_001042497
NM_133647
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr15_-_63199228
|
0.242
|
|
PDCD7
|
programmed cell death 7
|
|
chr12_-_51253875
|
0.241
|
NM_175053
|
KRT74
|
keratin 74
|
|
chr14_-_93924663
|
0.240
|
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr14_-_105610144
|
0.239
|
|
IGHA1
IGHG1
IGHG3
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
|
|
chr16_+_29734785
|
0.238
|
|
C16orf53
|
chromosome 16 open reading frame 53
|
|
chr10_-_104168890
|
0.237
|
NM_002779
|
PSD
|
pleckstrin and Sec7 domain containing
|
|
chrX_-_7855400
|
0.237
|
NM_004650
|
PNPLA4
|
patatin-like phospholipase domain containing 4
|
|
chr4_+_74493174
|
0.236
|
|
ALB
|
albumin
|
|
chr13_+_52114565
|
0.234
|
|
HNRNPA1L2
|
heterogeneous nuclear ribonucleoprotein A1-like 2
|
|
chr1_+_57093030
|
0.234
|
NM_000562
|
C8A
|
complement component 8, alpha polypeptide
|
|
chr4_+_74493218
|
0.234
|
|
ALB
|
albumin
|
|
chr1_-_27604656
|
0.230
|
|
WASF2
|
WAS protein family, member 2
|
|
chr19_+_61151009
|
0.230
|
NM_176811
|
NLRP8
|
NLR family, pyrin domain containing 8
|
|
chr18_-_29882555
|
0.230
|
NM_001198549
|
NOL4
|
nucleolar protein 4
|
|
chr1_+_240078655
|
0.229
|
NM_003686
NM_006027
|
EXO1
|
exonuclease 1
|
|
chr20_-_23915431
|
0.229
|
NM_178312
|
GGTLC1
|
gamma-glutamyltransferase light chain 1
|
|
chr2_+_88966170
|
0.229
|
|
|
|
|
chr1_+_173111306
|
0.228
|
NM_001035230
|
RABGAP1L
|
RAB GTPase activating protein 1-like
|
|
chr10_+_104169560
|
0.228
|
NM_024326
|
FBXL15
|
F-box and leucine-rich repeat protein 15
|
|
chr2_-_89073496
|
0.226
|
|
IGKC
|
immunoglobulin kappa constant
|
|
chr13_-_45654327
|
0.226
|
NM_002298
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr4_-_70539922
|
0.225
|
NM_001105677
|
UGT2A2
|
UDP glucuronosyltransferase 2 family, polypeptide A2
|
|
chr12_+_21059896
|
0.224
|
NM_001009562
|
LST-3TM12
|
organic anion transporter LST-3b
|
|
chr2_-_74235860
|
0.223
|
|
|
|
|
chr1_-_202726068
|
0.220
|
NM_002646
|
PIK3C2B
|
phosphoinositide-3-kinase, class 2, beta polypeptide
|
|
chr5_+_180399830
|
0.219
|
NM_152547
|
BTNL9
|
butyrophilin-like 9
|
|
chr4_+_74488910
|
0.219
|
|
ALB
|
albumin
|
|
chr9_+_33016482
|
0.219
|
|
DNAJA1
|
DnaJ (Hsp40) homolog, subfamily A, member 1
|
|
chr3_+_195336625
|
0.218
|
NM_005524
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr5_+_67558217
|
0.218
|
NM_181523
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr1_-_24342158
|
0.216
|
NM_021258
|
IL22RA1
|
interleukin 22 receptor, alpha 1
|
|
chr10_+_17891348
|
0.215
|
NM_001009567
NM_002438
|
MRC1L1
MRC1
|
mannose receptor, C type 1-like 1
mannose receptor, C type 1
|
|
chr14_+_94117483
|
0.214
|
NM_000624
|
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5
|
|
chr4_+_74496575
|
0.213
|
|
ALB
|
albumin
|
|
chr17_+_31562577
|
0.212
|
NM_207007
NM_001001435
|
CCL4L2
CCL4L1
|
chemokine (C-C motif) ligand 4-like 2
chemokine (C-C motif) ligand 4-like 1
|
|
chr18_-_56190980
|
0.211
|
NM_005912
|
MC4R
|
melanocortin 4 receptor
|
|
chr12_-_89922888
|
0.211
|
NM_004950
|
EPYC
|
epiphycan
|
|
chr8_+_22900863
|
0.208
|
NM_001160036
|
RHOBTB2
|
Rho-related BTB domain containing 2
|
|
chr12_+_51783554
|
0.206
|
|
SOAT2
|
sterol O-acyltransferase 2
|
|
chr10_-_116408046
|
0.203
|
NM_002313
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr3_+_48239865
|
0.202
|
NM_004345
|
CAMP
|
cathelicidin antimicrobial peptide
|
|
chr8_+_28230567
|
0.202
|
NM_006228
|
PNOC
|
prepronociceptin
|
|
chr6_-_119512035
|
0.202
|
NM_001100411
|
FAM184A
|
family with sequence similarity 184, member A
|
|
chr14_-_94677807
|
0.200
|
NM_030621
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr12_-_642786
|
0.200
|
NM_016533
|
NINJ2
|
ninjurin 2
|
|
chr12_+_119617060
|
0.200
|
|
|
|
|
chr5_-_150583808
|
0.197
|
|
CCDC69
|
coiled-coil domain containing 69
|
|
chr18_-_58005268
|
0.196
|
NM_012327
NM_176787
|
PIGN
|
phosphatidylinositol glycan anchor biosynthesis, class N
|
|
chr12_-_66933492
|
0.196
|
NM_020525
|
IL22
|
interleukin 22
|
|
chr3_-_137395966
|
0.195
|
NM_001145417
|
MSL2
|
male-specific lethal 2 homolog (Drosophila)
|
|
chr10_-_52315436
|
0.195
|
NM_001198818
NM_001198819
NM_001198820
NM_014576
NM_138932
NM_138933
|
A1CF
|
APOBEC1 complementation factor
|
|
chr17_+_43165608
|
0.195
|
NM_013351
|
TBX21
|
T-box 21
|
|
chr1_-_28392966
|
0.193
|
NM_001164722
NM_001164721
|
PTAFR
|
platelet-activating factor receptor
|
|
chr10_-_105835609
|
0.190
|
NM_000494
|
COL17A1
|
collagen, type XVII, alpha 1
|