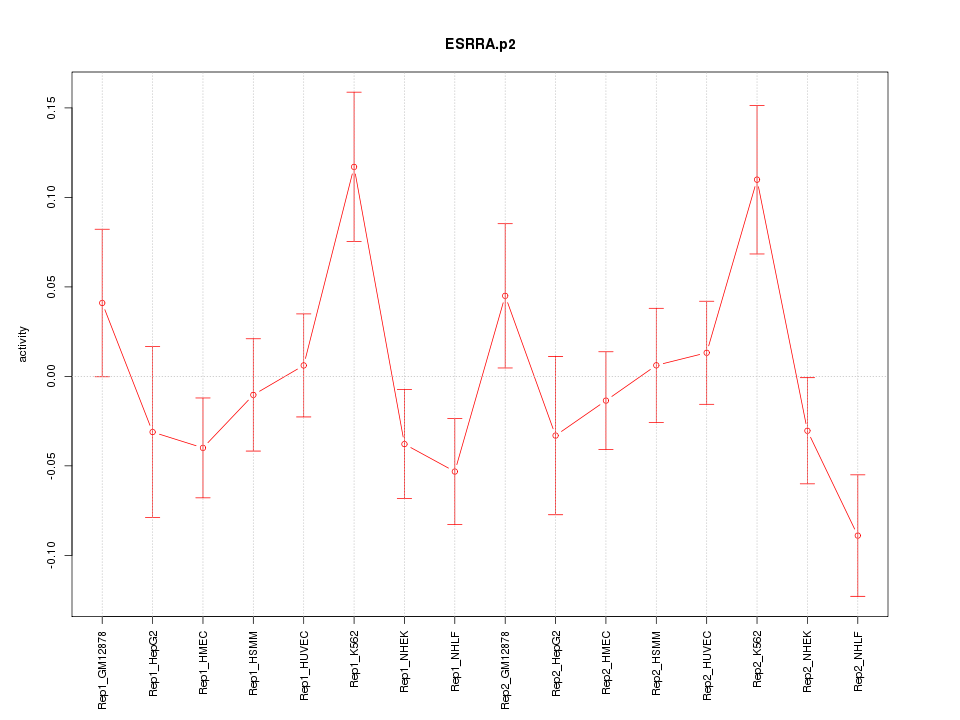
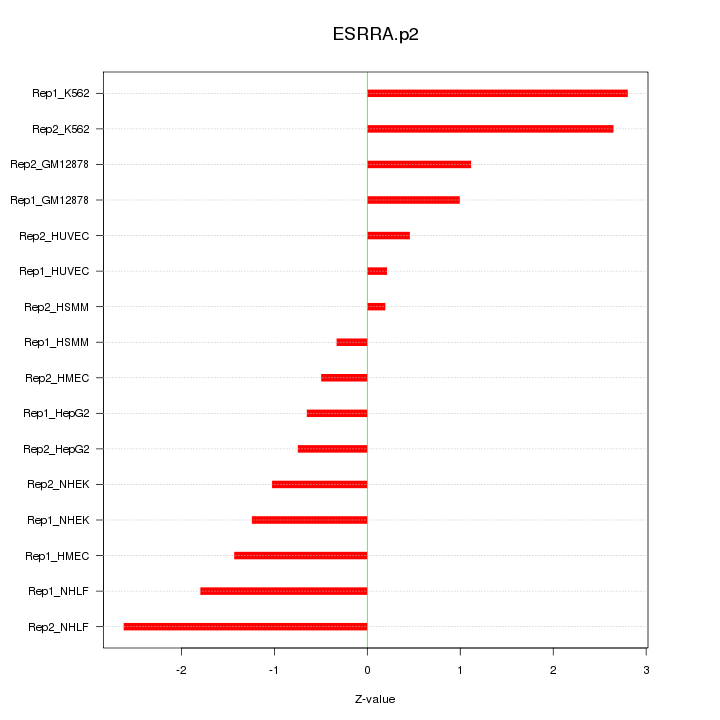
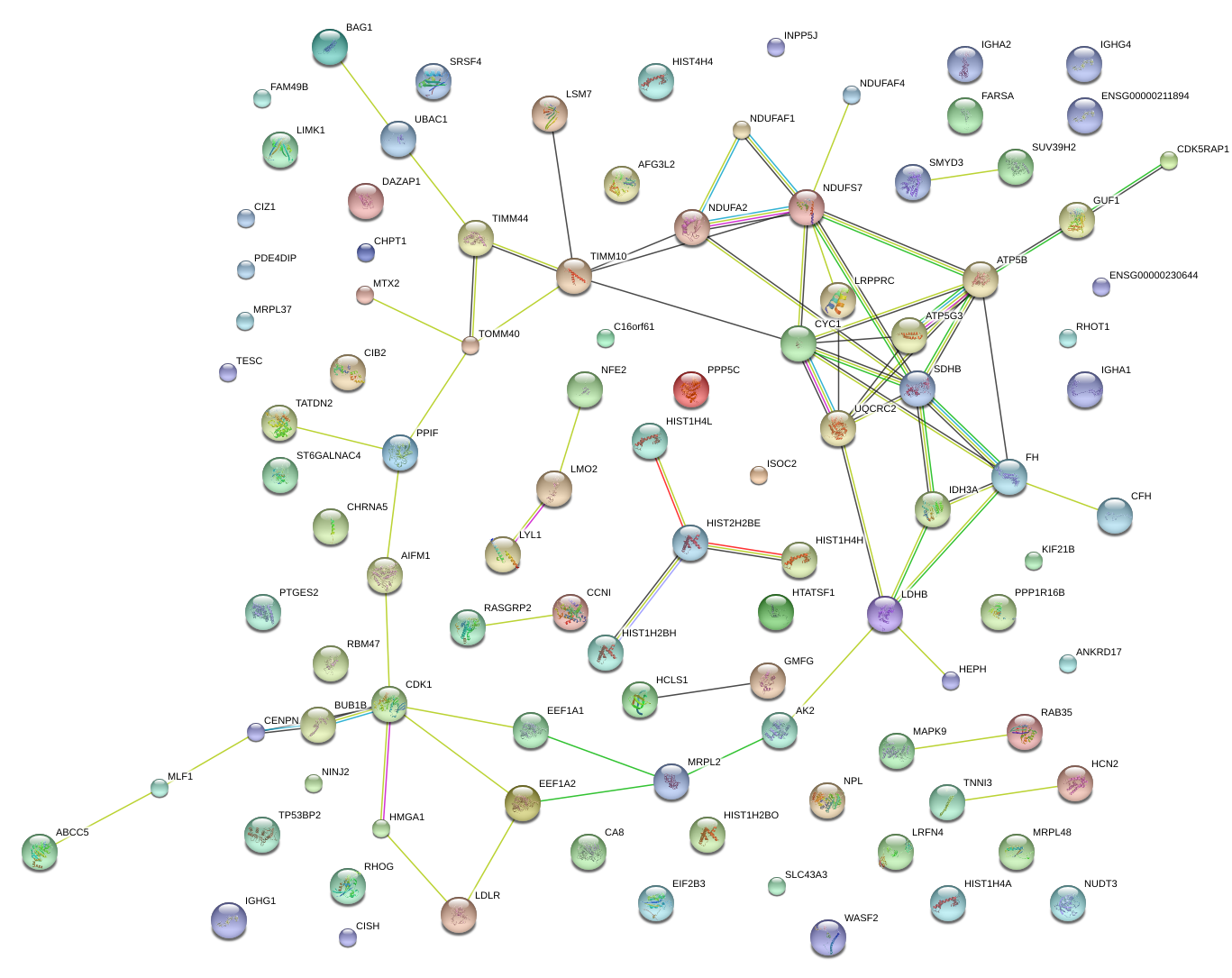
|
chr19_-_44518516
|
3.358
|
NM_004877
|
GMFG
|
glia maturation factor, gamma
|
|
chr19_-_44518465
|
2.915
|
|
GMFG
|
glia maturation factor, gamma
|
|
chr10_+_80777265
|
2.507
|
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr10_+_80777248
|
2.330
|
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr8_+_145221973
|
2.155
|
|
CYC1
|
cytochrome c-1
|
|
chr12_-_52980984
|
1.903
|
NM_001136023
|
NFE2
|
nuclear factor (erythroid-derived 2), 45kDa
|
|
chr20_+_36867708
|
1.881
|
NM_001172735
NM_015568
|
PPP1R16B
|
protein phosphatase 1, regulatory (inhibitor) subunit 16B
|
|
chr6_-_97452416
|
1.867
|
|
NDUFAF4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 4
|
|
chr6_-_26393705
|
1.820
|
NM_003543
|
HIST1H4H
|
histone cluster 1, H4h
|
|
chr12_-_52980914
|
1.811
|
|
NFE2
|
nuclear factor (erythroid-derived 2), 45kDa
|
|
chr8_-_131097856
|
1.691
|
|
FAM49B
|
family with sequence similarity 49, member B
|
|
chr3_-_50624085
|
1.554
|
|
CISH
|
cytokine inducible SH2-containing protein
|
|
chr11_-_56951167
|
1.421
|
NM_199329
|
SLC43A3
|
solute carrier family 43, member 3
|
|
chr11_-_56951132
|
1.383
|
|
SLC43A3
|
solute carrier family 43, member 3
|
|
chr19_-_60360768
|
1.380
|
NM_000363
|
TNNI3
|
troponin I type 3 (cardiac)
|
|
chr1_-_143706196
|
1.342
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr1_-_143706265
|
1.321
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr1_-_45224784
|
1.267
|
|
EIF2B3
|
eukaryotic translation initiation factor 2B, subunit 3 gamma, 58kDa
|
|
chr12_-_21701985
|
1.243
|
NM_001174097
|
LDHB
|
lactate dehydrogenase B
|
|
chr9_-_137992967
|
1.240
|
NM_016172
|
UBAC1
|
UBA domain containing 1
|
|
chr12_-_21702000
|
1.221
|
|
LDHB
|
lactate dehydrogenase B
|
|
chr1_-_33275071
|
1.216
|
NM_013411
|
AK2
|
adenylate kinase 2
|
|
chr12_-_21702036
|
1.213
|
|
LDHB
|
lactate dehydrogenase B
|
|
chr10_+_80777234
|
1.212
|
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr20_-_61600834
|
1.208
|
NM_001958
|
EEF1A2
|
eukaryotic translation elongation factor 1 alpha 2
|
|
chr12_-_21702021
|
1.189
|
|
LDHB
|
lactate dehydrogenase B
|
|
chr12_-_21702046
|
1.180
|
NM_002300
|
LDHB
|
lactate dehydrogenase B
|
|
chr8_-_131097828
|
1.154
|
|
FAM49B
|
family with sequence similarity 49, member B
|
|
chr10_+_80777199
|
1.141
|
NM_005729
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr14_-_105245959
|
1.137
|
|
IGHA1
|
immunoglobulin heavy constant alpha 1
|
|
chr1_-_33275015
|
1.118
|
|
AK2
|
adenylate kinase 2
|
|
chr1_-_17253073
|
1.107
|
NM_003000
|
SDHB
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip)
|
|
chr9_-_137992766
|
1.099
|
|
UBAC1
|
UBA domain containing 1
|
|
chr6_+_26359805
|
1.070
|
NM_003524
|
HIST1H2BH
|
histone cluster 1, H2bh
|
|
chr4_+_44375165
|
1.067
|
NM_021927
|
GUF1
|
GUF1 GTPase homolog (S. cerevisiae)
|
|
chr3_-_122862391
|
1.064
|
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr16_+_79598016
|
1.041
|
|
CENPN
|
centromere protein N
|
|
chr3_-_122862447
|
1.034
|
NM_005335
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr6_-_97452481
|
1.031
|
NM_014165
|
NDUFAF4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 4
|
|
chr11_-_56951088
|
1.028
|
NM_014096
|
SLC43A3
|
solute carrier family 43, member 3
|
|
chr8_+_145221924
|
1.011
|
NM_001916
|
CYC1
|
cytochrome c-1
|
|
chr6_-_97452468
|
0.999
|
|
NDUFAF4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 4
|
|
chr8_+_145221966
|
0.996
|
|
CYC1
|
cytochrome c-1
|
|
chr11_-_64268841
|
0.980
|
NM_001098670
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated)
|
|
chr15_-_76210611
|
0.975
|
NM_006383
|
CIB2
|
calcium and integrin binding family member 2
|
|
chr9_-_130006309
|
0.969
|
NM_001131015
NM_012127
|
CIZ1
|
CDKN1A interacting zinc finger protein 1
|
|
chr8_-_131098015
|
0.967
|
|
FAM49B
|
family with sequence similarity 49, member B
|
|
chr2_-_44076543
|
0.950
|
|
LRPPRC
|
leucine-rich PPR-motif containing
|
|
chr18_-_12367209
|
0.933
|
|
AFG3L2
|
AFG3 ATPase family gene 3-like 2 (S. cerevisiae)
|
|
chr1_-_222100006
|
0.901
|
|
TP53BP2
|
tumor protein p53 binding protein, 2
|
|
chr2_-_44076635
|
0.876
|
NM_133259
|
LRPPRC
|
leucine-rich PPR-motif containing
|
|
chr19_-_2279566
|
0.870
|
NM_016199
|
LSM7
|
LSM7 homolog, U6 small nuclear RNA associated (S. cerevisiae)
|
|
chr3_+_10265357
|
0.860
|
|
TATDN2
|
TatD DNase domain containing 2
|
|
chr17_+_27493680
|
0.859
|
|
RHOT1
|
ras homolog gene family, member T1
|
|
chr19_-_12905459
|
0.845
|
|
FARSA
|
phenylalanyl-tRNA synthetase, alpha subunit
|
|
chr4_-_40212670
|
0.840
|
NM_019027
|
RBM47
|
RNA binding motif protein 47
|
|
chr17_+_27493654
|
0.839
|
|
RHOT1
|
ras homolog gene family, member T1
|
|
chrX_-_129127361
|
0.839
|
|
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1
|
|
chr15_+_38240555
|
0.817
|
|
BUB1B
|
budding uninhibited by benzimidazoles 1 homolog beta (yeast)
|
|
chr2_-_44076616
|
0.814
|
|
LRPPRC
|
leucine-rich PPR-motif containing
|
|
chr1_-_244647333
|
0.814
|
NM_022743
|
SMYD3
|
SET and MYND domain containing 3
|
|
chr6_-_43135092
|
0.809
|
NM_015950
|
MRPL2
|
mitochondrial ribosomal protein L2
|
|
chr17_+_27493618
|
0.808
|
|
RHOT1
|
ras homolog gene family, member T1
|
|
chr1_-_239749683
|
0.808
|
NM_000143
|
FH
|
fumarate hydratase
|
|
chr19_-_60664860
|
0.802
|
NM_001136201
NM_001136202
NM_024710
|
ISOC2
|
isochorismatase domain containing 2
|
|
chr15_+_76228733
|
0.802
|
NM_005530
|
IDH3A
|
isocitrate dehydrogenase 3 (NAD+) alpha
|
|
chrX_-_129127310
|
0.795
|
|
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1
|
|
chr18_-_12367248
|
0.790
|
NM_006796
|
AFG3L2
|
AFG3 ATPase family gene 3-like 2 (S. cerevisiae)
|
|
chr15_+_76228792
|
0.787
|
|
IDH3A
|
isocitrate dehydrogenase 3 (NAD+) alpha
|
|
chr12_-_116021473
|
0.784
|
NM_001168325
NM_017899
|
TESC
|
tescalcin
|
|
chrX_+_65299157
|
0.778
|
NM_138737
|
HEPH
|
hephaestin
|
|
chr19_+_1358750
|
0.771
|
|
DAZAP1
|
DAZ associated protein 1
|
|
chr9_-_129719031
|
0.766
|
NM_175039
NM_175040
|
ST6GALNAC4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4
|
|
chr18_-_12367168
|
0.764
|
|
AFG3L2
|
AFG3 ATPase family gene 3-like 2 (S. cerevisiae)
|
|
chr19_-_12905524
|
0.763
|
|
FARSA
|
phenylalanyl-tRNA synthetase, alpha subunit
|
|
chrX_-_129127484
|
0.760
|
NM_145813
NM_001130847
NM_004208
NM_145812
|
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1
|
|
chr11_-_57054756
|
0.759
|
NM_012456
|
TIMM10
|
translocase of inner mitochondrial membrane 10 homolog (yeast)
|
|
chr12_-_642786
|
0.758
|
NM_016533
|
NINJ2
|
ninjurin 2
|
|
chr19_+_50086482
|
0.758
|
|
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast)
|
|
chr9_-_33254458
|
0.757
|
NM_001172415
NM_004323
|
BAG1
|
BCL2-associated athanogene
|
|
chr19_+_540849
|
0.747
|
NM_001194
|
HCN2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2
|
|
chr4_-_74307637
|
0.745
|
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chrX_+_135407336
|
0.743
|
NM_014500
|
HTATSF1
|
HIV-1 Tat specific factor 1
|
|
chr1_-_33275039
|
0.730
|
|
AK2
|
adenylate kinase 2
|
|
chr12_-_119038941
|
0.730
|
|
RAB35
|
RAB35, member RAS oncogene family
|
|
chr12_+_100615530
|
0.730
|
NM_020244
|
CHPT1
|
choline phosphotransferase 1
|
|
chr3_-_185218311
|
0.730
|
NM_001023587
NM_005688
|
ABCC5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5
|
|
chr1_-_33275057
|
0.724
|
NM_001625
|
AK2
|
adenylate kinase 2
|
|
chr5_-_140007351
|
0.723
|
|
NDUFA2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa
|
|
chr11_+_73176561
|
0.723
|
NM_016055
|
MRPL48
|
mitochondrial ribosomal protein L48
|
|
chr19_-_60664809
|
0.719
|
|
ISOC2
|
isochorismatase domain containing 2
|
|
chr2_-_175754366
|
0.719
|
NM_001002258
NM_001190329
NM_001689
|
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9)
|
|
chr19_-_13074973
|
0.718
|
NM_005583
|
LYL1
|
lymphoblastic leukemia derived sequence 1
|
|
chr19_-_12905528
|
0.716
|
NM_004461
|
FARSA
|
phenylalanyl-tRNA synthetase, alpha subunit
|
|
chr14_-_106084519
|
0.715
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr1_-_199259447
|
0.703
|
NM_017596
|
KIF21B
|
kinesin family member 21B
|
|
chr17_+_27493486
|
0.697
|
NM_001033566
NM_001033568
NM_018307
|
RHOT1
|
ras homolog gene family, member T1
|
|
chr16_+_21872201
|
0.697
|
|
UQCRC2
|
ubiquinol-cytochrome c reductase core protein II
|
|
chr5_-_179651569
|
0.696
|
|
MAPK9
|
mitogen-activated protein kinase 9
|
|
chr10_+_62208094
|
0.696
|
NM_001170406
NM_001170407
NM_001786
NM_033379
|
CDK1
|
cyclin-dependent kinase 1
|
|
chr1_-_29380945
|
0.695
|
|
SRSF4
|
serine/arginine-rich splicing factor 4
|
|
chr6_+_34314545
|
0.689
|
NM_145905
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr22_+_29848923
|
0.688
|
NM_001002837
|
INPP5J
|
inositol polyphosphate-5-phosphatase J
|
|
chr7_+_73136254
|
0.683
|
|
LIMK1
|
LIM domain kinase 1
|
|
chr12_-_55325912
|
0.681
|
|
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide
|
|
chr3_+_159771646
|
0.681
|
NM_001130156
NM_001130157
NM_001195432
NM_001195433
NM_001195434
NM_022443
|
MLF1
|
myeloid leukemia factor 1
|
|
chr16_-_79597940
|
0.673
|
NM_020188
|
C16orf61
|
chromosome 16 open reading frame 61
|
|
chr19_+_51542085
|
0.666
|
NM_006247
|
PPP5C
|
protein phosphatase 5, catalytic subunit
|
|
chr10_+_14960787
|
0.664
|
NM_001193424
NM_001193426
NM_001193425
NM_001193427
NM_024670
|
SUV39H2
|
suppressor of variegation 3-9 homolog 2 (Drosophila)
|
|
chr16_-_2663375
|
0.660
|
|
|
|
|
chr11_+_66381077
|
0.656
|
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4
|
|
chr16_+_21872109
|
0.653
|
NM_003366
|
UQCRC2
|
ubiquinol-cytochrome c reductase core protein II
|
|
chr16_+_21872155
|
0.650
|
|
UQCRC2
|
ubiquinol-cytochrome c reductase core protein II
|
|
chr1_-_33275051
|
0.647
|
|
AK2
|
adenylate kinase 2
|
|
chr16_+_21872178
|
0.638
|
|
UQCRC2
|
ubiquinol-cytochrome c reductase core protein II
|
|
chr15_+_76644882
|
0.637
|
NM_000745
|
CHRNA5
|
cholinergic receptor, nicotinic, alpha 5
|
|
chr2_+_176842368
|
0.637
|
NM_006554
|
MTX2
|
metaxin 2
|
|
chr19_-_7914510
|
0.636
|
|
TIMM44
|
translocase of inner mitochondrial membrane 44 homolog (yeast)
|
|
chr8_-_61356436
|
0.636
|
NM_004056
|
CA8
|
carbonic anhydrase VIII
|
|
chr11_-_64268126
|
0.635
|
NM_001098671
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated)
|
|
chr19_-_60664747
|
0.635
|
|
ISOC2
|
isochorismatase domain containing 2
|
|
chr1_+_181028016
|
0.632
|
NM_030769
|
NPL
|
N-acetylneuraminate pyruvate lyase (dihydrodipicolinate synthase)
|
|
chr11_-_33848013
|
0.628
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr20_-_31452964
|
0.621
|
|
CDK5RAP1
|
CDK5 regulatory subunit associated protein 1
|
|
chr11_-_3818713
|
0.621
|
|
RHOG
|
ras homolog gene family, member G (rho G)
|
|
chr11_-_33848061
|
0.619
|
NM_001142315
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr1_-_29381223
|
0.616
|
NM_005626
|
SRSF4
|
serine/arginine-rich splicing factor 4
|
|
chr19_+_1334898
|
0.611
|
|
NDUFS7
|
NADH dehydrogenase (ubiquinone) Fe-S protein 7, 20kDa (NADH-coenzyme Q reductase)
|
|
chr9_-_129929809
|
0.610
|
|
PTGES2
|
prostaglandin E synthase 2
|
|
chr1_-_27689254
|
0.605
|
NM_006990
|
WASF2
|
WAS protein family, member 2
|
|
chr6_-_34468076
|
0.602
|
|
NUDT3
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3
|
|
chr15_-_39481903
|
0.602
|
NM_016013
|
NDUFAF1
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 1
|
|
chr22_+_42726423
|
0.599
|
NM_001003828
|
PARVB
|
parvin, beta
|
|
chr17_+_77289804
|
0.598
|
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10
|
|
chr11_+_117777313
|
0.595
|
NM_006476
|
ATP5L
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G
|
|
chr10_+_22645304
|
0.592
|
NM_012071
|
COMMD3-BMI1
COMMD3
|
COMMD3-BMI1 read-through transcript
COMM domain containing 3
|
|
chr10_+_62208256
|
0.590
|
|
CDK1
|
cyclin-dependent kinase 1
|
|
chr19_-_7914605
|
0.589
|
NM_006351
|
TIMM44
|
translocase of inner mitochondrial membrane 44 homolog (yeast)
|
|
chr20_-_2589476
|
0.589
|
|
IDH3B
|
isocitrate dehydrogenase 3 (NAD+) beta
|
|
chr17_+_77289775
|
0.587
|
NM_012140
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10
|
|
chr1_-_27604656
|
0.574
|
|
WASF2
|
WAS protein family, member 2
|
|
chr9_+_130005489
|
0.572
|
|
DNM1
|
dynamin 1
|
|
chr6_-_34468288
|
0.570
|
|
NUDT3
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3
|
|
chr15_+_38520594
|
0.568
|
NM_014952
|
BAHD1
|
bromo adjacent homology domain containing 1
|
|
chr20_+_37024383
|
0.556
|
NM_001190809
NM_021931
|
DHX35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35
|
|
chr19_+_62434188
|
0.555
|
NM_001015879
NM_003160
NM_001015878
|
AURKC
|
aurora kinase C
|
|
chr22_+_18485068
|
0.555
|
|
RANBP1
|
RAN binding protein 1
|
|
chr16_+_79597552
|
0.551
|
NM_001100624
NM_001100625
NM_018455
|
CENPN
|
centromere protein N
|
|
chr19_+_1334904
|
0.549
|
|
NDUFS7
|
NADH dehydrogenase (ubiquinone) Fe-S protein 7, 20kDa (NADH-coenzyme Q reductase)
|
|
chr10_+_22645370
|
0.547
|
|
COMMD3
|
COMM domain containing 3
|
|
chr12_-_55326043
|
0.545
|
|
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide
|
|
chr17_-_26175764
|
0.544
|
|
CRLF3
|
cytokine receptor-like factor 3
|
|
chr19_+_1334877
|
0.543
|
NM_024407
|
NDUFS7
|
NADH dehydrogenase (ubiquinone) Fe-S protein 7, 20kDa (NADH-coenzyme Q reductase)
|
|
chr19_-_3013191
|
0.537
|
NM_001130
NM_198970
|
AES
|
amino-terminal enhancer of split
|
|
chr9_-_129929828
|
0.535
|
|
PTGES2
|
prostaglandin E synthase 2
|
|
chr1_+_200191241
|
0.534
|
NM_006335
|
TIMM17A
|
translocase of inner mitochondrial membrane 17 homolog A (yeast)
|
|
chr7_-_140270749
|
0.534
|
NM_004333
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B1
|
|
chr2_+_198073358
|
0.533
|
|
HSPE1
|
heat shock 10kDa protein 1 (chaperonin 10)
|
|
chr12_+_56462794
|
0.532
|
NM_001172695
NM_001172696
NM_001172697
NM_005726
|
TSFM
|
Ts translation elongation factor, mitochondrial
|
|
chr19_-_3013441
|
0.532
|
|
AES
|
amino-terminal enhancer of split
|
|
chr2_+_198073257
|
0.532
|
|
HSPE1
|
heat shock 10kDa protein 1 (chaperonin 10)
|
|
chr1_+_47572075
|
0.531
|
|
CMPK1
|
cytidine monophosphate (UMP-CMP) kinase 1, cytosolic
|
|
chr9_-_129929847
|
0.526
|
|
PTGES2
|
prostaglandin E synthase 2
|
|
chr1_-_29380850
|
0.525
|
|
SRSF4
|
serine/arginine-rich splicing factor 4
|
|
chr9_+_130005487
|
0.525
|
|
DNM1
|
dynamin 1
|
|
chr17_-_26175850
|
0.521
|
NM_015986
|
CRLF3
|
cytokine receptor-like factor 3
|
|
chr1_+_47572034
|
0.517
|
NM_001136140
NM_016308
|
CMPK1
|
cytidine monophosphate (UMP-CMP) kinase 1, cytosolic
|
|
chr12_-_52356375
|
0.511
|
NM_005176
|
ATP5G2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9)
|
|
chr19_-_4486202
|
0.509
|
NM_001013706
|
PLIN5
|
perilipin 5
|
|
chr9_+_130005429
|
0.508
|
NM_001005336
NM_004408
|
DNM1
|
dynamin 1
|
|
chr1_-_155053124
|
0.507
|
NM_001161441
NM_001161442
NM_001161444
NM_003975
|
SH2D2A
|
SH2 domain containing 2A
|
|
chr5_+_176807931
|
0.504
|
NM_001174102
|
PRR7
|
proline rich 7 (synaptic)
|
|
chr6_+_33280396
|
0.504
|
NM_014234
|
HSD17B8
|
hydroxysteroid (17-beta) dehydrogenase 8
|
|
chrX_+_118486419
|
0.503
|
|
SLC25A5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5
|
|
chr3_+_185520744
|
0.502
|
NM_004953
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr12_-_55326052
|
0.501
|
NM_001686
|
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide
|
|
chr3_+_157070933
|
0.490
|
NM_003875
|
GMPS
|
guanine monphosphate synthetase
|
|
chr9_-_129929765
|
0.488
|
|
PTGES2
|
prostaglandin E synthase 2
|
|
chr20_-_57051231
|
0.487
|
NM_016045
|
SLMO2
|
slowmo homolog 2 (Drosophila)
|
|
chr1_-_29380969
|
0.485
|
|
SRSF4
|
serine/arginine-rich splicing factor 4
|
|
chr4_-_7120656
|
0.484
|
NM_025196
|
GRPEL1
|
GrpE-like 1, mitochondrial (E. coli)
|
|
chr5_-_140007415
|
0.482
|
NM_001185012
NM_002488
|
NDUFA2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa
|
|
chr14_-_103058865
|
0.481
|
NM_001823
|
CKB
|
creatine kinase, brain
|
|
chr15_+_76228788
|
0.476
|
|
IDH3A
|
isocitrate dehydrogenase 3 (NAD+) alpha
|
|
chr14_-_106270497
|
0.472
|
|
IGHG3
|
immunoglobulin heavy constant gamma 3 (G3m marker)
|
|
chr13_-_41433066
|
0.470
|
|
KIAA0564
|
KIAA0564
|
|
chr10_-_74682247
|
0.468
|
NM_016065
|
MRPS16
|
mitochondrial ribosomal protein S16
|
|
chrX_-_151688850
|
0.468
|
NM_005362
|
MAGEA3
|
melanoma antigen family A, 3
|
|
chr12_-_119038934
|
0.465
|
|
RAB35
|
RAB35, member RAS oncogene family
|
|
chr5_-_137939216
|
0.464
|
NM_004134
|
HSPA9
|
heat shock 70kDa protein 9 (mortalin)
|
|
chr1_-_1679940
|
0.460
|
NM_001198995
|
NADK
|
NAD kinase
|
|
chr19_-_15096748
|
0.458
|
|
ILVBL
|
ilvB (bacterial acetolactate synthase)-like
|
|
chr19_-_3013768
|
0.458
|
|
AES
|
amino-terminal enhancer of split
|
|
chr2_-_26320967
|
0.455
|
|
HADHA
|
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit
|
|
chr9_-_99994704
|
0.453
|
NM_052820
|
CORO2A
|
coronin, actin binding protein, 2A
|
|
chr11_-_75769514
|
0.449
|
NM_004705
|
PRKRIR
|
protein-kinase, interferon-inducible double stranded RNA dependent inhibitor, repressor of (P58 repressor)
|
|
chr16_+_79597621
|
0.448
|
|
CENPN
|
centromere protein N
|
|
chr12_-_52356123
|
0.446
|
|
ATP5G2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9)
|
|
chr1_+_32530289
|
0.445
|
|
HDAC1
|
histone deacetylase 1
|
|
chr2_-_198072805
|
0.435
|
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin)
|