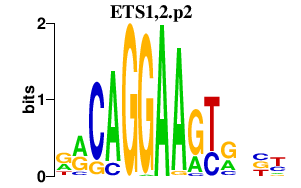
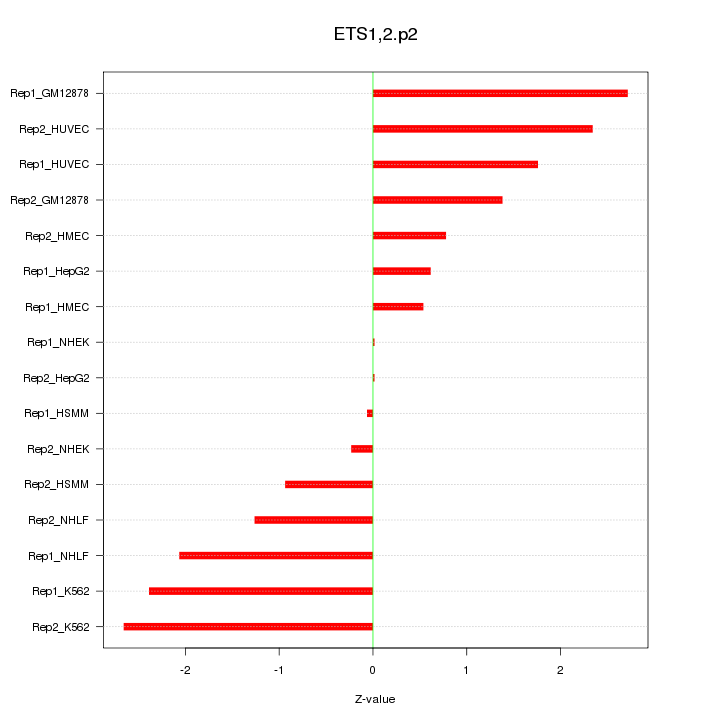
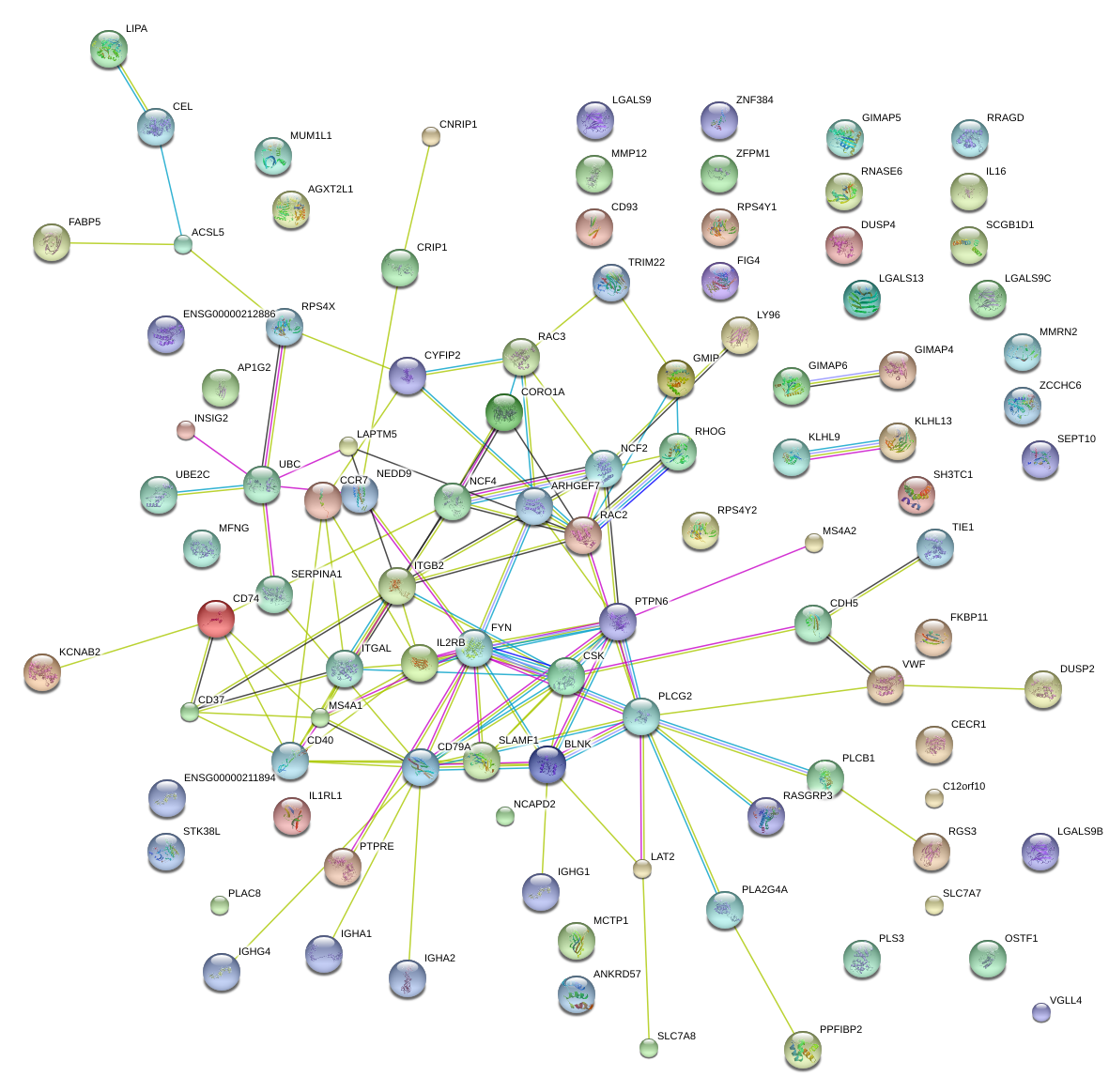
|
chr1_-_31003260
|
2.770
|
NM_006762
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chr1_-_31003181
|
2.575
|
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chr2_-_109728958
|
2.118
|
NM_144710
NM_178584
|
SEPT10
|
septin 10
|
|
chr19_+_54530546
|
2.050
|
|
CD37
|
CD37 molecule
|
|
chr2_+_33514896
|
1.890
|
NM_170672
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr12_-_47604930
|
1.838
|
NM_001143781
|
FKBP11
|
FK506 binding protein 11, 19 kDa
|
|
chr12_-_6103945
|
1.803
|
NM_000552
|
VWF
|
von Willebrand factor
|
|
chr9_+_115396020
|
1.756
|
NM_144489
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr19_+_54530512
|
1.679
|
|
CD37
|
CD37 molecule
|
|
chr10_-_98021144
|
1.676
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr19_+_54530497
|
1.632
|
|
CD37
|
CD37 molecule
|
|
chr19_+_54530485
|
1.631
|
NM_001040031
NM_001774
|
CD37
|
CD37 molecule
|
|
chr2_-_109728693
|
1.625
|
|
SEPT10
|
septin 10
|
|
chr21_-_45165249
|
1.519
|
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit)
|
|
chr6_-_11490487
|
1.496
|
NM_001142393
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr19_+_54530458
|
1.482
|
|
CD37
|
CD37 molecule
|
|
chr21_-_45165302
|
1.479
|
NM_000211
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit)
|
|
chr5_-_149772469
|
1.442
|
NM_001025159
NM_004355
NM_001025158
|
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain
|
|
chr14_-_106184988
|
1.414
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr7_-_149960612
|
1.377
|
NM_024711
|
GIMAP6
|
GTPase, IMAP family member 6
|
|
chr7_-_149960218
|
1.359
|
|
GIMAP6
|
GTPase, IMAP family member 6
|
|
chr5_-_149772563
|
1.354
|
|
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain
|
|
chr10_-_88707299
|
1.320
|
NM_024756
|
MMRN2
|
multimerin 2
|
|
chr14_+_20319049
|
1.296
|
NM_005615
|
RNASE6
|
ribonuclease, RNase A family, k6
|
|
chr16_+_30102426
|
1.281
|
|
CORO1A
|
coronin, actin binding protein, 1A
|
|
chr2_-_96174865
|
1.240
|
NM_004418
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr12_+_6925997
|
1.238
|
NM_080548
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr12_+_6926014
|
1.223
|
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr8_+_75066117
|
1.217
|
NM_001195797
NM_015364
|
LY96
|
lymphocyte antigen 96
|
|
chr1_-_181826086
|
1.192
|
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr22_-_35875890
|
1.186
|
NM_000878
|
IL2RB
|
interleukin 2 receptor, beta
|
|
chr10_+_129595289
|
1.162
|
NM_006504
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr21_-_45165204
|
1.159
|
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit)
|
|
chr11_+_5667494
|
1.129
|
NM_006074
|
TRIM22
|
tripartite motif containing 22
|
|
chr20_+_43874735
|
1.078
|
|
UBE2C
|
ubiquitin-conjugating enzyme E2C
|
|
chr2_+_118562499
|
1.076
|
NM_016133
|
INSIG2
|
insulin induced gene 2
|
|
chrX_+_114734074
|
1.073
|
NM_001136025
NM_001172335
|
PLS3
|
plastin 3
|
|
chr9_-_88159143
|
1.050
|
NM_001185059
NM_001185074
NM_024617
|
ZCCHC6
|
zinc finger, CCHC domain containing 6
|
|
chr14_+_105024576
|
1.042
|
|
CRIP1
|
cysteine-rich protein 1 (intestinal)
|
|
chr14_-_22354457
|
1.020
|
|
SLC7A7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7
|
|
chr11_+_7574992
|
1.010
|
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr2_+_102320148
|
0.998
|
NM_003856
|
IL1RL1
|
interleukin 1 receptor-like 1
|
|
chr4_+_8251818
|
0.968
|
NM_018986
|
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1
|
|
chr11_+_59979857
|
0.940
|
NM_021950
NM_152866
|
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1
|
|
chr14_-_93926679
|
0.934
|
NM_001002235
NM_001002236
NM_001127700
NM_001127701
NM_001127702
NM_001127703
NM_001127704
NM_001127705
NM_001127706
NM_001127707
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr5_+_156625629
|
0.918
|
NM_001037333
NM_001037332
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr20_+_43874654
|
0.916
|
NM_007019
NM_181799
NM_181800
NM_181802
NM_181803
|
UBE2C
|
ubiquitin-conjugating enzyme E2C
|
|
chr7_+_149895390
|
0.916
|
NM_018326
|
GIMAP4
|
GTPase, IMAP family member 4
|
|
chr1_-_181826292
|
0.902
|
NM_000433
NM_001190789
NM_001190794
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr12_+_27288344
|
0.884
|
NM_015000
|
STK38L
|
serine/threonine kinase 38 like
|
|
chr12_+_51979721
|
0.883
|
NM_021640
|
C12orf10
|
chromosome 12 open reading frame 10
|
|
chr13_+_110604039
|
0.883
|
NM_003899
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7
|
|
chr5_-_94443300
|
0.872
|
NM_001002796
|
MCTP1
|
multiple C2 domains, transmembrane 1
|
|
chr17_+_18320822
|
0.870
|
NM_001040078
|
LGALS9B
LGALS9C
|
lectin, galactoside-binding, soluble, 9B
lectin, galactoside-binding, soluble, 9C
|
|
chr17_-_35975227
|
0.866
|
NM_001838
|
CCR7
|
chemokine (C-C motif) receptor 7
|
|
chrY_+_2769596
|
0.864
|
NM_001008
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1
|
|
chrY_+_2769630
|
0.861
|
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1
|
|
chr1_+_43539239
|
0.856
|
NM_005424
|
TIE1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1
|
|
chr6_-_90178630
|
0.842
|
|
RRAGD
|
Ras-related GTP binding D
|
|
chr16_+_30391460
|
0.839
|
NM_001114380
NM_002209
|
ITGAL
|
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide)
|
|
chr7_+_150065376
|
0.838
|
NM_018384
|
GIMAP5
|
GTPase, IMAP family member 5
|
|
chr6_+_110119190
|
0.831
|
|
FIG4
|
FIG4 homolog, SAC1 lipid phosphatase domain containing (S. cerevisiae)
|
|
chr20_+_43875078
|
0.815
|
NM_181801
|
UBE2C
|
ubiquitin-conjugating enzyme E2C
|
|
chr22_+_35586950
|
0.809
|
NM_000631
NM_013416
|
NCF4
|
neutrophil cytosolic factor 4, 40kDa
|
|
chr4_-_84254873
|
0.796
|
|
PLAC8
|
placenta-specific 8
|
|
chr10_-_91092410
|
0.796
|
|
LIPA
|
lipase A, lysosomal acid, cholesterol esterase
|
|
chr16_+_80370426
|
0.796
|
NM_002661
|
PLCG2
|
phospholipase C, gamma 2 (phosphatidylinositol-specific)
|
|
chr14_-_23106372
|
0.794
|
|
AP1G2
|
adaptor-related protein complex 1, gamma 2 subunit
|
|
chr3_-_11660395
|
0.794
|
NM_001128219
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr1_+_6008956
|
0.791
|
NM_003636
NM_172130
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr8_+_82355322
|
0.791
|
NM_001444
|
FABP5
|
fatty acid binding protein 5 (psoriasis-associated)
|
|
chr4_-_84254912
|
0.790
|
|
PLAC8
|
placenta-specific 8
|
|
chr20_-_23014828
|
0.782
|
NM_012072
|
CD93
|
CD93 molecule
|
|
chr15_+_79376295
|
0.782
|
NM_004513
|
IL16
|
interleukin 16 (lymphocyte chemoattractant factor)
|
|
chr6_+_110119116
|
0.777
|
NM_014845
|
FIG4
|
FIG4 homolog, SAC1 lipid phosphatase domain containing (S. cerevisiae)
|
|
chr6_-_112186977
|
0.770
|
NM_153048
|
FYN
|
FYN oncogene related to SRC, FGR, YES
|
|
chr9_-_21325239
|
0.753
|
NM_018847
|
KLHL9
|
kelch-like 9 (Drosophila)
|
|
chr16_+_64958025
|
0.749
|
NM_001795
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr6_+_110119214
|
0.748
|
|
FIG4
|
FIG4 homolog, SAC1 lipid phosphatase domain containing (S. cerevisiae)
|
|
chr22_-_16060485
|
0.747
|
NM_177405
|
CECR1
|
cat eye syndrome chromosome region, candidate 1
|
|
chr17_+_22982319
|
0.741
|
|
LGALS9
|
lectin, galactoside-binding, soluble, 9
|
|
chr20_+_44180296
|
0.741
|
NM_001250
NM_152854
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5
|
|
chr1_-_158883666
|
0.729
|
NM_003037
|
SLAMF1
|
signaling lymphocytic activation molecule family member 1
|
|
chr14_-_105762742
|
0.728
|
|
|
|
|
chr12_+_6472782
|
0.726
|
|
NCAPD2
|
non-SMC condensin I complex, subunit D2
|
|
chr12_-_6668787
|
0.720
|
NM_001039918
NM_133476
|
ZNF384
|
zinc finger protein 384
|
|
chr22_-_36212160
|
0.716
|
NM_001166343
NM_002405
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr19_+_47073029
|
0.711
|
NM_001783
NM_021601
|
CD79A
|
CD79a molecule, immunoglobulin-associated alpha
|
|
chr17_+_22982344
|
0.710
|
|
LGALS9
|
lectin, galactoside-binding, soluble, 9
|
|
chr12_+_6930799
|
0.710
|
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr15_+_72861470
|
0.708
|
NM_001127190
NM_004383
|
CSK
|
c-src tyrosine kinase
|
|
chr1_+_185064654
|
0.706
|
NM_024420
|
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent)
|
|
chr2_+_109729176
|
0.699
|
NM_023016
|
ANKRD57
|
ankyrin repeat domain 57
|
|
chr9_+_76893232
|
0.698
|
|
OSTF1
|
osteoclast stimulating factor 1
|
|
chr12_-_47605478
|
0.696
|
NM_001143782
NM_016594
|
FKBP11
|
FK506 binding protein 11, 19 kDa
|
|
chr22_-_35970133
|
0.694
|
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr10_+_114125955
|
0.694
|
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr8_-_29262240
|
0.693
|
NM_057158
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr12_+_6930778
|
0.691
|
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr22_-_35970230
|
0.685
|
NM_002872
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr14_-_22354851
|
0.684
|
NM_001126105
NM_003982
|
SLC7A7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7
|
|
chr12_+_6930815
|
0.682
|
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr7_+_73262283
|
0.680
|
|
LAT2
|
linker for activation of T cells family, member 2
|
|
chr7_+_150065594
|
0.677
|
|
GIMAP5
|
GTPase, IMAP family member 5
|
|
chr12_+_6930694
|
0.666
|
NM_002831
NM_080549
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr19_-_19615452
|
0.663
|
NM_016573
|
GMIP
|
GEM interacting protein
|
|
chr3_-_126325132
|
0.654
|
|
SLC12A8
|
solute carrier family 12 (potassium/chloride transporters), member 8
|
|
chrX_+_128741562
|
0.648
|
NM_018990
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr7_+_73262022
|
0.645
|
NM_014146
NM_032463
NM_032464
|
LAT2
|
linker for activation of T cells family, member 2
|
|
chr16_+_30391550
|
0.645
|
|
ITGAL
|
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide)
|
|
chr9_-_122716670
|
0.644
|
NM_001190947
|
TRAF1
|
TNF receptor-associated factor 1
|
|
chr3_-_122862447
|
0.643
|
NM_005335
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr12_+_6930792
|
0.643
|
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr1_+_32489426
|
0.643
|
NM_005356
|
LCK
|
lymphocyte-specific protein tyrosine kinase
|
|
chr6_-_29635456
|
0.638
|
|
UBD
|
ubiquitin D
|
|
chr14_+_23771942
|
0.638
|
NM_001002002
NM_016576
|
GMPR2
|
guanosine monophosphate reductase 2
|
|
chr17_+_4784097
|
0.637
|
|
RNF167
|
ring finger protein 167
|
|
chr17_-_20311411
|
0.637
|
NM_001042685
|
LGALS9B
|
lectin, galactoside-binding, soluble, 9B
|
|
chrX_+_128741578
|
0.635
|
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr12_+_6930804
|
0.631
|
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr10_+_26767657
|
0.630
|
|
APBB1IP
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein
|
|
chr10_+_114126004
|
0.629
|
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr12_-_91063441
|
0.627
|
NM_001731
|
BTG1
|
B-cell translocation gene 1, anti-proliferative
|
|
chr6_-_90178685
|
0.626
|
NM_021244
|
RRAGD
|
Ras-related GTP binding D
|
|
chr3_-_173010921
|
0.625
|
NM_001130081
NM_002662
|
PLD1
|
phospholipase D1, phosphatidylcholine-specific
|
|
chr15_-_68971665
|
0.621
|
NM_020147
|
THAP10
|
THAP domain containing 10
|
|
chr12_-_109372489
|
0.620
|
|
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa
|
|
chr14_+_105024301
|
0.617
|
NM_001311
|
CRIP1
|
cysteine-rich protein 1 (intestinal)
|
|
chr10_+_26767698
|
0.617
|
|
APBB1IP
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein
|
|
chr3_-_122862391
|
0.609
|
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr4_+_2783764
|
0.609
|
|
SH3BP2
|
SH3-domain binding protein 2
|
|
chr17_+_4784524
|
0.602
|
|
RNF167
|
ring finger protein 167
|
|
chr1_+_32512298
|
0.600
|
NM_001042771
|
LCK
|
lymphocyte-specific protein tyrosine kinase
|
|
chr14_+_75687997
|
0.599
|
NM_017926
NM_017972
|
C14orf118
|
chromosome 14 open reading frame 118
|
|
chr1_-_27825178
|
0.594
|
NM_001042747
|
FGR
|
Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog
|
|
chr17_+_4784330
|
0.590
|
NM_015528
|
RNF167
|
ring finger protein 167
|
|
chr21_+_17807167
|
0.589
|
NM_001338
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr6_-_90178544
|
0.589
|
|
RRAGD
|
Ras-related GTP binding D
|
|
chr7_+_73262264
|
0.587
|
|
LAT2
|
linker for activation of T cells family, member 2
|
|
chr14_+_75688042
|
0.583
|
|
C14orf118
|
chromosome 14 open reading frame 118
|
|
chr8_-_101391356
|
0.582
|
|
RNF19A
|
ring finger protein 19A
|
|
chr22_+_21053972
|
0.579
|
|
|
|
|
chr7_-_5535726
|
0.578
|
|
ACTB
|
actin, beta
|
|
chr12_-_109372481
|
0.575
|
|
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa
|
|
chr17_+_22982372
|
0.575
|
|
LGALS9
|
lectin, galactoside-binding, soluble, 9
|
|
chr6_-_32892705
|
0.573
|
NM_002120
|
HLA-DOB
|
major histocompatibility complex, class II, DO beta
|
|
chr12_-_109372535
|
0.563
|
NM_005719
|
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa
|
|
chr2_-_175170674
|
0.563
|
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr16_+_80370400
|
0.561
|
|
PLCG2
|
phospholipase C, gamma 2 (phosphatidylinositol-specific)
|
|
chr20_+_44180355
|
0.557
|
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5
|
|
chr11_-_88864061
|
0.557
|
NM_001143836
NM_016931
|
NOX4
|
NADPH oxidase 4
|
|
chr18_+_13208777
|
0.556
|
NM_181481
NM_181482
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr12_-_6668880
|
0.556
|
NM_001039917
NM_001039919
NM_001039920
NM_001135734
|
ZNF384
|
zinc finger protein 384
|
|
chr2_+_30223304
|
0.553
|
|
YPEL5
|
yippee-like 5 (Drosophila)
|
|
chr2_+_30223372
|
0.553
|
|
YPEL5
|
yippee-like 5 (Drosophila)
|
|
chr5_-_151046638
|
0.553
|
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr5_+_134012133
|
0.553
|
NM_021982
|
SEC24A
|
SEC24 family, member A (S. cerevisiae)
|
|
chrX_-_106905657
|
0.552
|
NM_198057
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr6_-_32605898
|
0.552
|
NM_002125
|
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 5
|
|
chr4_+_68107039
|
0.548
|
NM_012108
|
STAP1
|
signal transducing adaptor family member 1
|
|
chr1_+_181380837
|
0.547
|
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2)
|
|
chr2_+_30223253
|
0.546
|
NM_001127399
NM_001127400
NM_001127401
NM_016061
|
YPEL5
|
yippee-like 5 (Drosophila)
|
|
chr8_+_56954899
|
0.540
|
NM_001111097
NM_002350
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
|
|
chr9_-_129656581
|
0.535
|
|
ENG
|
endoglin
|
|
chr9_+_127549437
|
0.534
|
NM_006195
|
PBX3
|
pre-B-cell leukemia homeobox 3
|
|
chr17_-_959007
|
0.532
|
NM_001092
|
ABR
|
active BCR-related gene
|
|
chr7_-_120285245
|
0.532
|
|
TSPAN12
|
tetraspanin 12
|
|
chr9_-_122728173
|
0.529
|
|
TRAF1
|
TNF receptor-associated factor 1
|
|
chr17_+_22982278
|
0.528
|
NM_002308
NM_009587
|
LGALS9
|
lectin, galactoside-binding, soluble, 9
|
|
chr3_+_48482213
|
0.527
|
NM_016381
NM_033629
|
TREX1
|
three prime repair exonuclease 1
|
|
chr9_-_129656830
|
0.527
|
NM_000118
NM_001114753
|
ENG
|
endoglin
|
|
chr17_+_29606408
|
0.526
|
NM_002982
|
CCL2
|
chemokine (C-C motif) ligand 2
|
|
chr2_+_218790109
|
0.526
|
NM_152862
|
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa
|
|
chr8_+_56954946
|
0.525
|
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
|
|
chr10_+_129735796
|
0.525
|
NM_130435
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr2_-_69724209
|
0.525
|
|
AAK1
|
AP2 associated kinase 1
|
|
chr6_-_133126278
|
0.522
|
NM_078488
|
VNN2
|
vanin 2
|
|
chr9_+_115382760
|
0.520
|
NM_021106
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr12_-_51887318
|
0.517
|
|
ITGB7
|
integrin, beta 7
|
|
chr9_-_129656778
|
0.516
|
|
ENG
|
endoglin
|
|
chr15_-_78050515
|
0.515
|
NM_001114735
NM_004049
|
BCL2A1
|
BCL2-related protein A1
|
|
chr14_-_76993641
|
0.514
|
NM_001193314
NM_001193315
NM_001193316
NM_001193317
NM_022067
|
VIPAR
|
VPS33B interacting protein, apical-basolateral polarity regulator
|
|
chr9_-_129656413
|
0.513
|
|
ENG
|
endoglin
|
|
chr19_-_19615369
|
0.511
|
|
GMIP
|
GEM interacting protein
|
|
chr1_-_24067347
|
0.511
|
NM_000147
|
FUCA1
|
fucosidase, alpha-L- 1, tissue
|
|
chr5_-_171547700
|
0.508
|
|
STK10
|
serine/threonine kinase 10
|
|
chr19_-_43800374
|
0.508
|
NM_001042600
NM_007181
|
MAP4K1
|
mitogen-activated protein kinase kinase kinase kinase 1
|
|
chr6_-_32665409
|
0.508
|
NM_002124
|
HLA-DRB1
HLA-DRB4
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 1
major histocompatibility complex, class II, DR beta 4
major histocompatibility complex, class II, DR beta 5
|
|
chr11_-_66961709
|
0.505
|
NM_005608
|
PTPRCAP
|
protein tyrosine phosphatase, receptor type, C-associated protein
|
|
chr3_+_120799411
|
0.505
|
NM_015900
|
PLA1A
|
phospholipase A1 member A
|
|
chr1_-_205161821
|
0.503
|
NM_001193338
NM_001142473
NM_005449
|
FAIM3
|
Fas apoptotic inhibitory molecule 3
|
|
chr2_+_71148915
|
0.501
|
NM_017567
|
NAGK
|
N-acetylglucosamine kinase
|
|
chr12_+_64504408
|
0.501
|
NM_003483
NM_003484
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr12_-_109372462
|
0.499
|
|
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa
|
|
chr9_-_76892860
|
0.499
|
NM_001127603
NM_017881
|
C9orf95
|
chromosome 9 open reading frame 95
|
|
chr2_+_218790143
|
0.498
|
|
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa
|
|
chr8_+_38763878
|
0.496
|
NM_001122824
NM_006283
|
TACC1
|
transforming, acidic coiled-coil containing protein 1
|
|
chr14_-_105644300
|
0.494
|
|
IGHV4-31
IGHA1
IGHA2
IGHG1
IGHG3
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
|
|
chr3_-_11736946
|
0.491
|
NM_014667
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr2_+_230798687
|
0.491
|
NM_001005176
NM_007237
|
SP140
|
SP140 nuclear body protein
|