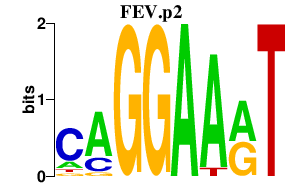
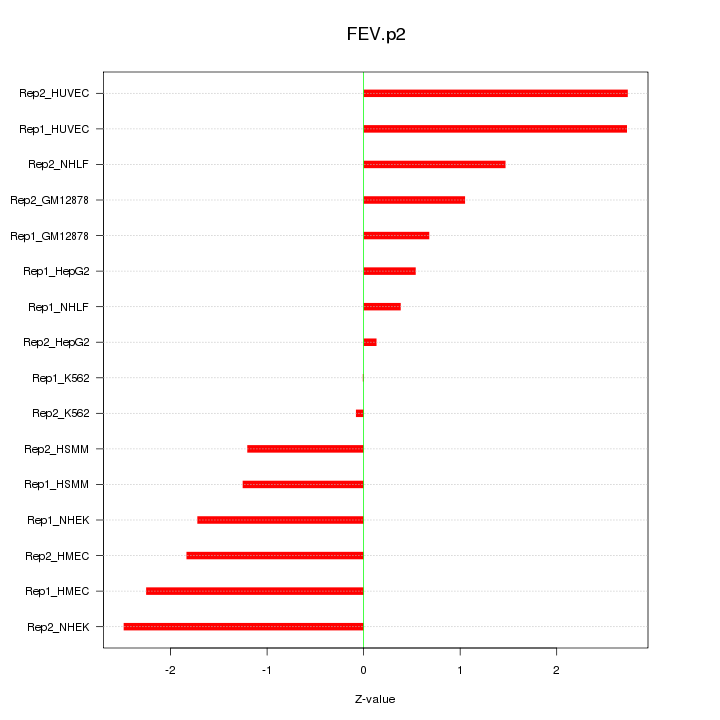
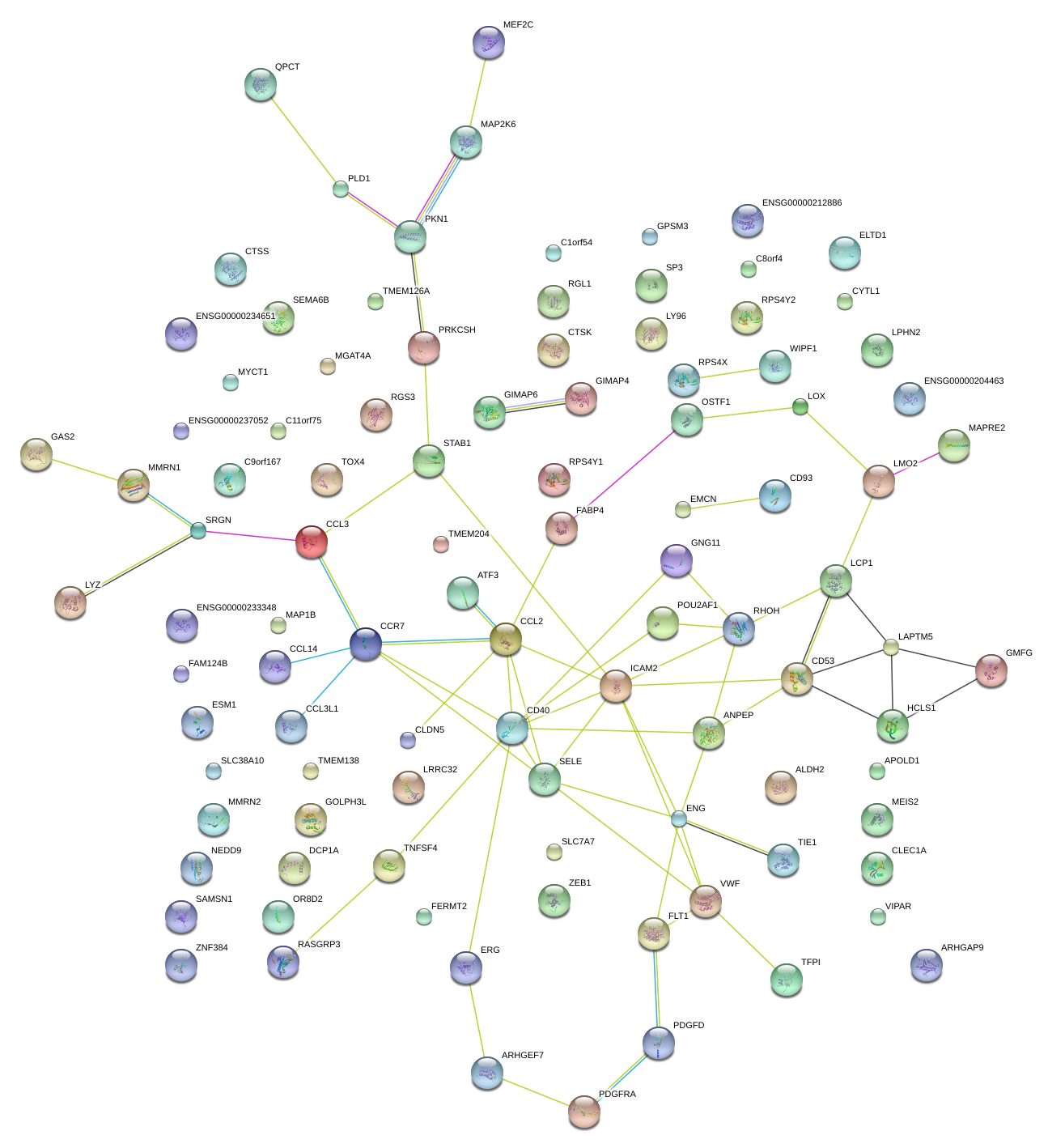
|
chr3_-_122862447
|
3.071
|
NM_005335
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr3_-_122862391
|
2.913
|
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr8_-_82557956
|
2.157
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr19_-_44518465
|
1.971
|
|
GMFG
|
glia maturation factor, gamma
|
|
chr17_-_59437782
|
1.907
|
NM_000873
|
ICAM2
|
intercellular adhesion molecule 2
|
|
chr1_-_79244948
|
1.894
|
NM_022159
|
ELTD1
|
EGF, latrophilin and seven transmembrane domain containing 1
|
|
chr19_-_44518516
|
1.865
|
NM_004877
|
GMFG
|
glia maturation factor, gamma
|
|
chr2_+_33514896
|
1.849
|
NM_170672
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr4_-_101658094
|
1.648
|
NM_001159694
NM_016242
|
EMCN
|
endomucin
|
|
chr12_-_6103945
|
1.633
|
NM_000552
|
VWF
|
von Willebrand factor
|
|
chr5_-_54317102
|
1.513
|
NM_001135604
NM_007036
|
ESM1
|
endothelial cell-specific molecule 1
|
|
chrY_+_2769630
|
1.310
|
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1
|
|
chr11_-_76059435
|
1.305
|
NM_001128922
|
LRRC32
|
leucine rich repeat containing 32
|
|
chrY_+_2769596
|
1.274
|
NM_001008
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1
|
|
chr10_+_31650069
|
1.221
|
NM_001128128
NM_001174094
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr16_+_1523658
|
1.191
|
|
TMEM204
|
transmembrane protein 204
|
|
chr4_+_91035025
|
1.185
|
NM_007351
|
MMRN1
|
multimerin 1
|
|
chr21_-_14840506
|
1.124
|
NM_022136
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1
|
|
chr8_+_75066117
|
1.113
|
NM_001195797
NM_015364
|
LY96
|
lymphocyte antigen 96
|
|
chr17_-_59451662
|
1.080
|
NM_001099786
NM_001099787
NM_001099788
NM_001099789
|
ICAM2
|
intercellular adhesion molecule 2
|
|
chr11_-_33848061
|
1.074
|
NM_001142315
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr1_-_31003260
|
1.017
|
NM_006762
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chr11_-_33848013
|
1.001
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr6_-_11490487
|
0.997
|
NM_001142393
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr2_-_175207521
|
0.985
|
NM_003387
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr9_-_129656830
|
0.938
|
NM_000118
NM_001114753
|
ENG
|
endoglin
|
|
chr11_-_33847834
|
0.931
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr11_-_33847937
|
0.921
|
NM_001142316
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr21_-_38792173
|
0.893
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr9_-_129656778
|
0.890
|
|
ENG
|
endoglin
|
|
chr8_+_40130143
|
0.882
|
NM_020130
|
C8orf4
|
chromosome 8 open reading frame 4
|
|
chr22_-_17892859
|
0.857
|
NM_001130861
NM_003277
|
CLDN5
|
claudin 5
|
|
chr14_-_22354851
|
0.847
|
NM_001126105
NM_003982
|
SLC7A7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7
|
|
chr1_+_43539239
|
0.844
|
NM_005424
|
TIE1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1
|
|
chr9_-_129656581
|
0.839
|
|
ENG
|
endoglin
|
|
chr5_+_71538456
|
0.823
|
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr11_-_92916161
|
0.813
|
NM_020179
|
C11orf75
|
chromosome 11 open reading frame 75
|
|
chr7_+_93388951
|
0.793
|
NM_004126
|
GNG11
|
guanine nucleotide binding protein (G protein), gamma 11
|
|
chr11_-_33847507
|
0.788
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr11_-_103539895
|
0.785
|
|
PDGFD
|
platelet derived growth factor D
|
|
chr9_+_115396020
|
0.763
|
NM_144489
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr11_-_103540275
|
0.742
|
|
PDGFD
|
platelet derived growth factor D
|
|
chr6_-_11340869
|
0.739
|
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr11_-_103540233
|
0.715
|
NM_025208
NM_033135
|
PDGFD
|
platelet derived growth factor D
|
|
chr6_+_153060722
|
0.710
|
NM_025107
|
MYCT1
|
myc target 1
|
|
chr4_+_54790020
|
0.707
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr7_-_149960612
|
0.700
|
NM_024711
|
GIMAP6
|
GTPase, IMAP family member 6
|
|
chr17_+_64922420
|
0.695
|
NM_002758
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr12_+_68028407
|
0.694
|
|
LYZ
|
lysozyme
|
|
chr11_-_110755112
|
0.681
|
|
POU2AF1
|
POU class 2 associating factor 1
|
|
chr11_+_22644732
|
0.677
|
NM_001143830
|
GAS2
|
growth arrest-specific 2
|
|
chr17_+_29606408
|
0.673
|
NM_002982
|
CCL2
|
chemokine (C-C motif) ligand 2
|
|
chr17_-_59817722
|
0.659
|
NM_000442
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr10_+_70517863
|
0.640
|
|
SRGN
|
serglycin
|
|
chr1_-_149047329
|
0.639
|
NM_000396
|
CTSK
|
cathepsin K
|
|
chr13_+_110604039
|
0.633
|
NM_003899
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7
|
|
chr10_+_70517823
|
0.626
|
NM_002727
|
SRGN
|
serglycin
|
|
chr13_+_110604071
|
0.618
|
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7
|
|
chr3_+_52504391
|
0.617
|
NM_015136
|
STAB1
|
stabilin 1
|
|
chr12_-_6668680
|
0.614
|
|
ZNF384
|
zinc finger protein 384
|
|
chr4_+_39874921
|
0.610
|
NM_004310
|
RHOH
|
ras homolog gene family, member H
|
|
chr10_-_88707299
|
0.600
|
NM_024756
|
MMRN2
|
multimerin 2
|
|
chr9_-_129656413
|
0.598
|
|
ENG
|
endoglin
|
|
chr1_+_182040837
|
0.594
|
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr14_-_76993641
|
0.593
|
NM_001193314
NM_001193315
NM_001193316
NM_001193317
NM_022067
|
VIPAR
|
VPS33B interacting protein, apical-basolateral polarity regulator
|
|
chr4_-_5071996
|
0.586
|
NM_018659
|
CYTL1
|
cytokine-like 1
|
|
chr6_-_32268267
|
0.584
|
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr15_-_35179614
|
0.578
|
|
MEIS2
|
Meis homeobox 2
|
|
chr7_+_149895390
|
0.574
|
NM_018326
|
GIMAP4
|
GTPase, IMAP family member 4
|
|
chr13_-_45654296
|
0.566
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr18_+_30875297
|
0.564
|
NM_014268
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr1_-_167969802
|
0.559
|
NM_000450
|
SELE
|
selectin E
|
|
chr5_-_121440704
|
0.557
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr12_+_12829807
|
0.557
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr3_-_173010921
|
0.556
|
NM_001130081
NM_002662
|
PLD1
|
phospholipase D1, phosphatidylcholine-specific
|
|
chr1_+_210805319
|
0.555
|
NM_001030287
|
ATF3
|
activating transcription factor 3
|
|
chr13_-_27967215
|
0.548
|
NM_001159920
NM_001160030
NM_001160031
NM_002019
|
FLT1
|
fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor)
|
|
chr17_-_31441390
|
0.547
|
NM_002983
|
CCL3
|
chemokine (C-C motif) ligand 3
|
|
chr13_-_45654327
|
0.540
|
NM_002298
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr1_-_149004879
|
0.536
|
|
CTSS
|
cathepsin S
|
|
chr17_-_59817654
|
0.535
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr3_+_52504410
|
0.534
|
|
STAB1
|
stabilin 1
|
|
chr6_-_11340887
|
0.533
|
NM_006403
NM_182966
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr19_+_14412087
|
0.531
|
|
PKN1
|
protein kinase N1
|
|
chr17_-_35975227
|
0.520
|
NM_001838
|
CCR7
|
chemokine (C-C motif) receptor 7
|
|
chr12_-_10142779
|
0.516
|
NM_016511
|
CLEC1A
|
C-type lectin domain family 1, member A
|
|
chr11_-_110755366
|
0.514
|
NM_006235
|
POU2AF1
|
POU class 2 associating factor 1
|
|
chr1_-_148936123
|
0.513
|
NM_018178
|
GOLPH3L
|
golgi phosphoprotein 3-like
|
|
chr1_+_82038669
|
0.511
|
NM_012302
|
LPHN2
|
latrophilin 2
|
|
chr2_+_37425240
|
0.507
|
NM_012413
|
QPCT
|
glutaminyl-peptide cyclotransferase
|
|
chr12_+_110688728
|
0.505
|
NM_000690
|
ALDH2
|
aldehyde dehydrogenase 2 family (mitochondrial)
|
|
chr17_-_59817456
|
0.501
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr9_+_139292094
|
0.499
|
NM_017723
|
C9orf167
|
chromosome 9 open reading frame 167
|
|
chr2_-_98646367
|
0.498
|
NM_001160154
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr11_+_60886407
|
0.498
|
|
TMEM138
|
transmembrane protein 138
|
|
chr19_-_4510713
|
0.496
|
|
SEMA6B
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B
|
|
chr20_-_23014828
|
0.496
|
NM_012072
|
CD93
|
CD93 molecule
|
|
chr3_-_53356573
|
0.494
|
NM_018403
|
DCP1A
|
DCP1 decapping enzyme homolog A (S. cerevisiae)
|
|
chr1_-_171441234
|
0.492
|
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr9_+_76893232
|
0.491
|
|
OSTF1
|
osteoclast stimulating factor 1
|
|
chr17_-_76883677
|
0.490
|
NM_001037984
NM_138570
|
SLC38A10
|
solute carrier family 38, member 10
|
|
chr20_+_44180296
|
0.489
|
NM_001250
NM_152854
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5
|
|
chr6_-_32268615
|
0.487
|
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr1_+_148511806
|
0.483
|
NM_024579
|
C1orf54
|
chromosome 1 open reading frame 54
|
|
chr5_-_88235624
|
0.482
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr2_-_188127430
|
0.480
|
NM_001032281
NM_006287
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr6_-_31728405
|
0.477
|
|
BAG6
|
BCL2-associated athanogene 6
|
|
chr17_-_59817575
|
0.476
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr17_-_31337798
|
0.472
|
NM_032962
NM_032963
|
CCL14
|
chemokine (C-C motif) ligand 14
|
|
chr2_-_174538197
|
0.471
|
|
SP3
|
Sp3 transcription factor
|
|
chr2_-_224974954
|
0.469
|
NM_001122779
NM_024785
|
FAM124B
|
family with sequence similarity 124B
|
|
chr9_+_115367122
|
0.465
|
NM_134427
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr7_-_149960218
|
0.463
|
|
GIMAP6
|
GTPase, IMAP family member 6
|
|
chr5_-_88214720
|
0.463
|
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr15_-_88148177
|
0.461
|
|
ANPEP
|
alanyl (membrane) aminopeptidase
|
|
chr1_-_31003181
|
0.459
|
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chr14_-_52487386
|
0.457
|
|
FERMT2
|
fermitin family member 2
|
|
chr1_+_111217244
|
0.450
|
NM_000560
|
CD53
|
CD53 molecule
|
|
chr14_-_53491019
|
0.448
|
NM_130851
|
BMP4
|
bone morphogenetic protein 4
|
|
chr4_-_16509354
|
0.448
|
|
LDB2
|
LIM domain binding 2
|
|
chr4_-_16509446
|
0.447
|
NM_001130834
NM_001290
|
LDB2
|
LIM domain binding 2
|
|
chr16_+_29373365
|
0.447
|
NM_001014999
NM_001015000
NM_024044
NM_178044
|
SLX1A-SULT1A3
SLX1A
SLX1B
|
SLX1A-SULT1A3 read-through transcript
SLX1 structure-specific endonuclease subunit homolog A (S. cerevisiae)
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae)
|
|
chr16_+_64958025
|
0.444
|
NM_001795
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr6_-_31728363
|
0.439
|
|
BAG6
|
BCL2-associated athanogene 6
|
|
chr5_+_95092384
|
0.436
|
NM_014899
|
RHOBTB3
|
Rho-related BTB domain containing 3
|
|
chr13_+_57103944
|
0.434
|
|
PCDH17
|
protocadherin 17
|
|
chr2_-_136590210
|
0.434
|
NM_001008540
|
CXCR4
|
chemokine (C-X-C motif) receptor 4
|
|
chr1_+_111217314
|
0.433
|
|
CD53
|
CD53 molecule
|
|
chr8_+_38704860
|
0.430
|
NM_001146216
|
TACC1
|
transforming, acidic coiled-coil containing protein 1
|
|
chr6_+_107917965
|
0.429
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr4_-_16509126
|
0.427
|
|
LDB2
|
LIM domain binding 2
|
|
chr3_+_134976965
|
0.426
|
|
TF
|
transferrin
|
|
chr1_+_196874839
|
0.424
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr6_-_7855125
|
0.423
|
NM_001145549
|
TXNDC5
|
thioredoxin domain containing 5 (endoplasmic reticulum)
|
|
chr11_-_404975
|
0.422
|
NM_001135054
|
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain
|
|
chr1_-_36562341
|
0.415
|
NM_018166
|
FAM176B
|
family with sequence similarity 176, member B
|
|
chr15_+_79376295
|
0.415
|
NM_004513
|
IL16
|
interleukin 16 (lymphocyte chemoattractant factor)
|
|
chr1_-_149004852
|
0.414
|
|
CTSS
|
cathepsin S
|
|
chr11_-_88864061
|
0.414
|
NM_001143836
NM_016931
|
NOX4
|
NADPH oxidase 4
|
|
chr13_+_30089829
|
0.414
|
NM_005800
|
USPL1
|
ubiquitin specific peptidase like 1
|
|
chr16_+_75782608
|
0.413
|
|
MON1B
|
MON1 homolog B (yeast)
|
|
chr12_-_9123126
|
0.412
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr22_+_27498679
|
0.412
|
|
CCDC117
|
coiled-coil domain containing 117
|
|
chr7_-_115458064
|
0.408
|
|
TFEC
|
transcription factor EC
|
|
chr1_+_32512298
|
0.407
|
NM_001042771
|
LCK
|
lymphocyte-specific protein tyrosine kinase
|
|
chrX_+_12795111
|
0.403
|
NM_016562
|
TLR7
|
toll-like receptor 7
|
|
chr14_-_52487558
|
0.401
|
NM_001134999
NM_001135000
NM_006832
|
FERMT2
|
fermitin family member 2
|
|
chr5_-_93472993
|
0.399
|
NM_001163417
NM_001163418
NM_032042
|
FAM172A
|
family with sequence similarity 172, member A
|
|
chr1_-_161439248
|
0.397
|
NM_001195303
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr7_-_115458010
|
0.395
|
NM_001018058
NM_012252
|
TFEC
|
transcription factor EC
|
|
chr15_-_35179650
|
0.388
|
NM_170674
NM_170675
NM_170676
NM_170677
|
MEIS2
|
Meis homeobox 2
|
|
chr12_+_68028397
|
0.387
|
NM_000239
|
LYZ
|
lysozyme
|
|
chr6_-_112682573
|
0.386
|
|
LAMA4
|
laminin, alpha 4
|
|
chr19_-_10311262
|
0.386
|
NM_002162
|
ICAM3
|
intercellular adhesion molecule 3
|
|
chr6_-_31728447
|
0.385
|
NM_004639
NM_080702
NM_080703
|
BAG6
|
BCL2-associated athanogene 6
|
|
chr11_+_64620183
|
0.383
|
NM_013265
|
C11orf2
|
chromosome 11 open reading frame 2
|
|
chr11_-_124175493
|
0.382
|
NM_024631
|
C11orf61
|
chromosome 11 open reading frame 61
|
|
chr3_-_184756101
|
0.382
|
NM_130446
|
KLHL6
|
kelch-like 6 (Drosophila)
|
|
chr6_-_31728121
|
0.382
|
NM_001098534
|
BAG6
|
BCL2-associated athanogene 6
|
|
chr3_-_46224791
|
0.381
|
NM_001295
|
CCR1
|
chemokine (C-C motif) receptor 1
|
|
chr12_-_91063441
|
0.381
|
NM_001731
|
BTG1
|
B-cell translocation gene 1, anti-proliferative
|
|
chr1_+_196874759
|
0.381
|
NM_002838
NM_080921
NM_080923
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr11_+_64620276
|
0.380
|
|
C11orf2
|
chromosome 11 open reading frame 2
|
|
chr17_-_53950191
|
0.377
|
NM_004687
|
MTMR4
|
myotubularin related protein 4
|
|
chr10_+_114123765
|
0.377
|
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr3_+_11242666
|
0.374
|
NM_001098211
|
HRH1
|
histamine receptor H1
|
|
chr11_+_64620287
|
0.373
|
|
C11orf2
|
chromosome 11 open reading frame 2
|
|
chrX_+_15428820
|
0.370
|
NM_203281
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr10_+_114123828
|
0.369
|
NM_203379
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr14_+_20319049
|
0.367
|
NM_005615
|
RNASE6
|
ribonuclease, RNase A family, k6
|
|
chr17_-_26665144
|
0.365
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr7_-_135312734
|
0.363
|
NM_145808
NM_001128619
|
MTPN
LUZP6
|
myotrophin
leucine zipper protein 6
|
|
chrX_-_44945025
|
0.363
|
NM_024689
NM_176819
|
CXorf36
|
chromosome X open reading frame 36
|
|
chrX_-_15783015
|
0.362
|
NM_003916
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr2_-_188127294
|
0.361
|
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr5_-_121441910
|
0.359
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr4_+_86615275
|
0.358
|
NM_001025616
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr1_-_111544784
|
0.358
|
NM_024901
|
DENND2D
|
DENN/MADD domain containing 2D
|
|
chr5_-_149515613
|
0.358
|
NM_002609
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr5_-_111121150
|
0.357
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chrX_-_135691168
|
0.357
|
NM_004840
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr19_+_54530546
|
0.357
|
|
CD37
|
CD37 molecule
|
|
chr6_+_122834760
|
0.355
|
NM_181794
|
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta
|
|
chr2_+_102320148
|
0.354
|
NM_003856
|
IL1RL1
|
interleukin 1 receptor-like 1
|
|
chr17_+_37694007
|
0.353
|
|
STAT5A
|
signal transducer and activator of transcription 5A
|
|
chr17_-_26665220
|
0.351
|
NM_006495
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr5_-_149515331
|
0.350
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr19_+_52940610
|
0.349
|
|
GLTSCR2
|
glioma tumor suppressor candidate region gene 2
|
|
chr18_-_51219880
|
0.349
|
|
TCF4
|
transcription factor 4
|
|
chr14_-_22354457
|
0.347
|
|
SLC7A7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7
|
|
chr19_-_40928131
|
0.347
|
NM_001040425
NM_144987
|
U2AF1L4
|
U2 small nuclear RNA auxiliary factor 1-like 4
|
|
chr4_-_70661018
|
0.347
|
NM_014465
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1
|
|
chr17_-_44042924
|
0.345
|
|
HOXB7
|
homeobox B7
|
|
chrX_+_128741562
|
0.344
|
NM_018990
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr11_-_74818685
|
0.343
|
|
KLHL35
|
kelch-like 35 (Drosophila)
|
|
chr6_-_47030492
|
0.342
|
NM_015234
|
GPR116
|
G protein-coupled receptor 116
|
|
chr19_-_17377383
|
0.337
|
NM_004335
|
BST2
|
bone marrow stromal cell antigen 2
|
|
chr9_+_102244005
|
0.335
|
NM_001198812
|
C9orf3-TMEFF1
C9orf30
|
C9orf3-TMEFF1 readthrough
chromosome 9 open reading frame 30
|
|
chr16_-_24850163
|
0.334
|
|
ARHGAP17
|
Rho GTPase activating protein 17
|
|
chr20_+_44180355
|
0.332
|
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5
|