|
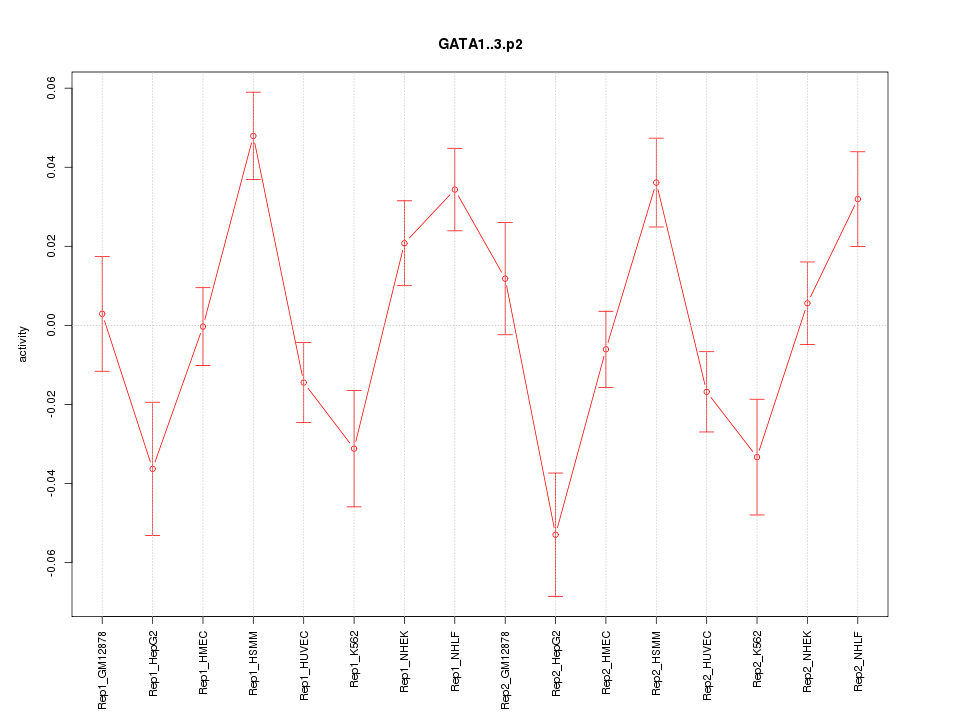
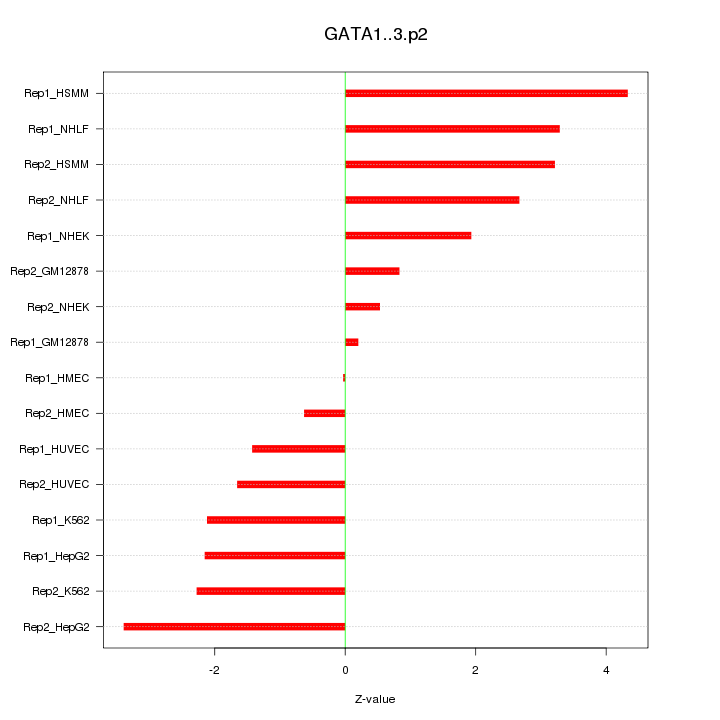
chr2_-_189752575
|
5.803
|
NM_000393
|
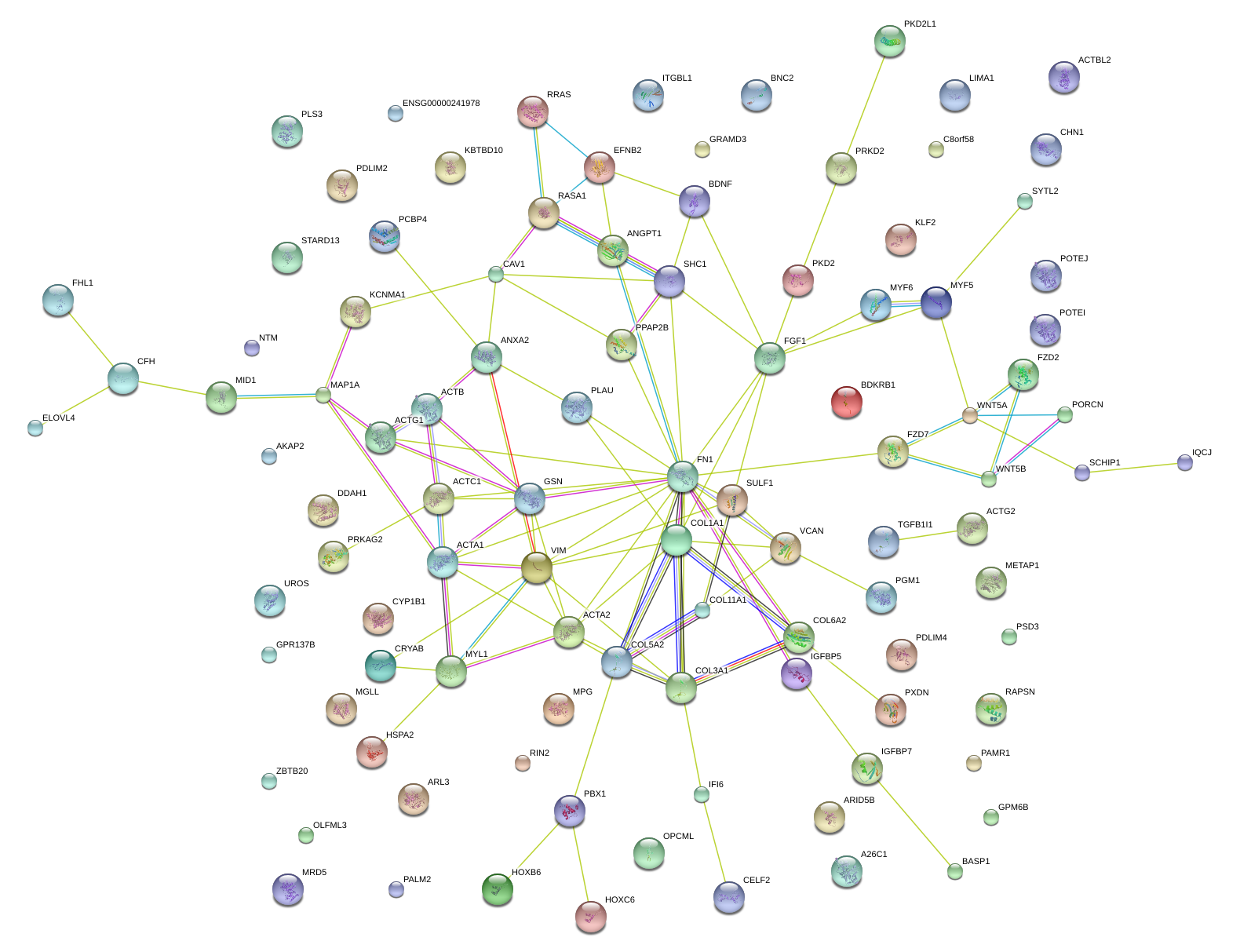
COL5A2
|
collagen, type V, alpha 2
|
|
chr12_+_1608672
|
4.949
|
NM_032642
|
WNT5B
|
wingless-type MMTV integration site family, member 5B
|
|
chr5_+_140870881
|
4.821
|
|
|
|
|
chr5_+_140871107
|
4.655
|
|
|
|
|
chr17_+_39990124
|
4.624
|
NM_001466
|
FZD2
|
frizzled homolog 2 (Drosophila)
|
|
chr11_-_35503720
|
4.501
|
NM_001001991
NM_015430
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr2_+_189547323
|
4.280
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr2_+_189547358
|
4.202
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr2_+_189547377
|
3.926
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr17_-_45617899
|
3.816
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr3_-_129023850
|
3.503
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr10_+_75343352
|
3.278
|
|
PLAU
|
plasminogen activator, urokinase
|
|
chr1_+_114323552
|
3.155
|
NM_020190
|
OLFML3
|
olfactomedin-like 3
|
|
chr3_+_161040343
|
2.790
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr17_-_45633873
|
2.753
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr9_-_16717887
|
2.741
|
|
BNC2
|
basonuclin 2
|
|
chr17_-_45619038
|
2.696
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr12_-_48902692
|
2.571
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr4_+_89147821
|
2.484
|
NM_000297
|
PKD2
|
polycystic kidney disease 2 (autosomal dominant)
|
|
chr12_+_79634820
|
2.356
|
NM_005593
|
MYF5
|
myogenic factor 5
|
|
chr1_-_27871287
|
2.300
|
|
IFI6
|
interferon, alpha-inducible protein 6
|
|
chr14_+_64077171
|
2.260
|
|
HSPA2
|
heat shock 70kDa protein 2
|
|
chr2_-_175578156
|
2.209
|
NM_001025201
NM_001822
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr11_-_111287657
|
2.140
|
NM_001885
|
CRYAB
|
crystallin, alpha B
|
|
chr11_+_130745580
|
2.059
|
NM_001048209
|
NTM
|
neurotrimin
|
|
chr1_-_27871245
|
2.058
|
|
IFI6
|
interferon, alpha-inducible protein 6
|
|
chr11_-_27698800
|
2.013
|
NM_001143805
NM_001143806
NM_170732
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr12_+_79625538
|
1.999
|
NM_002469
|
MYF6
|
myogenic factor 6 (herculin)
|
|
chr1_+_162795328
|
1.973
|
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr2_-_210876560
|
1.935
|
NM_079422
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast
|
|
chr3_+_161040439
|
1.898
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr2_-_215949566
|
1.863
|
|
FN1
|
fibronectin 1
|
|
chr8_+_70541412
|
1.811
|
NM_001128204
NM_015170
|
SULF1
|
sulfatase 1
|
|
chr8_+_70567581
|
1.800
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr1_-_103346482
|
1.794
|
|
COL11A1
|
collagen, type XI, alpha 1
|
|
chrX_-_10495476
|
1.791
|
NM_001193278
NM_001193279
NM_001193280
NM_001193281
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chrX_+_135058402
|
1.768
|
NM_001159700
|
FHL1
|
four and a half LIM domains 1
|
|
chr8_+_22493928
|
1.766
|
NM_176871
NM_198042
|
PDLIM2
|
PDZ and LIM domain 2 (mystique)
|
|
chr8_-_18915443
|
1.754
|
NM_015310
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr20_+_19863716
|
1.749
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr2_+_202607554
|
1.735
|
NM_003507
|
FZD7
|
frizzled homolog 7 (Drosophila)
|
|
chr3_-_55496607
|
1.725
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr7_-_5534737
|
1.680
|
|
ACTB
|
actin, beta
|
|
chr1_-_85816520
|
1.671
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr15_-_32875040
|
1.655
|
NM_005159
|
ACTC1
|
actin, alpha, cardiac muscle 1
|
|
chr8_+_70567634
|
1.645
|
|
SULF1
|
sulfatase 1
|
|
chr5_+_82803248
|
1.644
|
NM_001126336
NM_001164097
NM_001164098
NM_004385
|
VCAN
|
versican
|
|
chr2_+_170074457
|
1.607
|
NM_006063
|
KBTBD10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr13_+_100902942
|
1.601
|
NM_004791
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains)
|
|
chr10_+_17311242
|
1.594
|
|
VIM
|
vimentin
|
|
chr19_+_16296632
|
1.580
|
NM_016270
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr9_+_123126652
|
1.580
|
|
GSN
|
gelsolin
|
|
chr3_-_51968995
|
1.563
|
|
PCBP4
|
poly(rC) binding protein 4
|
|
chr10_+_17311347
|
1.558
|
|
VIM
|
vimentin
|
|
chr3_-_115826481
|
1.551
|
NM_001164346
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr7_-_151142858
|
1.546
|
NM_001040633
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit
|
|
chr10_+_17311307
|
1.544
|
|
VIM
|
vimentin
|
|
chr10_+_11247059
|
1.537
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr1_-_103346639
|
1.536
|
NM_001190709
NM_001854
NM_080629
NM_080630
|
COL11A1
|
collagen, type XI, alpha 1
|
|
chrX_+_48253007
|
1.534
|
NM_203474
NM_203475
|
PORCN
|
porcupine homolog (Drosophila)
|
|
chr9_+_111582317
|
1.525
|
NM_053016
NM_007203
NM_147150
|
PALM2
PALM2-AKAP2
|
paralemmin 2
PALM2-AKAP2 readthrough
|
|
chr11_-_85146691
|
1.507
|
NM_001162951
NM_001162953
|
SYTL2
|
synaptotagmin-like 2
|
|
chr13_-_105985313
|
1.490
|
NM_004093
|
EFNB2
|
ephrin-B2
|
|
chr1_-_56750235
|
1.477
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr10_-_79067552
|
1.477
|
NM_001014797
NM_001161352
NM_001161353
NM_002247
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chr15_+_41597097
|
1.450
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr8_-_18585721
|
1.447
|
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr2_-_217248967
|
1.440
|
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr10_+_17311282
|
1.431
|
|
VIM
|
vimentin
|
|
chr10_+_17311283
|
1.428
|
|
VIM
|
vimentin
|
|
chr11_-_47427263
|
1.422
|
NM_005055
NM_032645
|
RAPSN
|
receptor-associated protein of the synapse
|
|
chr19_-_54835162
|
1.421
|
NM_006270
|
RRAS
|
related RAS viral (r-ras) oncogene homolog
|
|
chr9_+_33614214
|
1.406
|
|
ANXA2P2
|
annexin A2 pseudogene 2
|
|
chr1_+_63861474
|
1.399
|
NM_001172818
|
PGM1
|
phosphoglucomutase 1
|
|
chr7_+_115952279
|
1.391
|
NM_001172895
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr5_+_125828685
|
1.382
|
NM_001146322
|
GRAMD3
|
GRAM domain containing 3
|
|
chr7_+_115952325
|
1.381
|
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr7_-_5534252
|
1.376
|
|
ACTB
|
actin, beta
|
|
chr14_+_95792299
|
1.362
|
NM_000710
|
BDKRB1
|
bradykinin receptor B1
|
|
chr21_+_46370220
|
1.360
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr5_+_131621287
|
1.356
|
|
PDLIM4
|
PDZ and LIM domain 4
|
|
chr7_+_115952074
|
1.330
|
NM_001753
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr10_+_63331018
|
1.298
|
NM_032199
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chrX_+_114701432
|
1.290
|
NM_005032
|
PLS3
|
plastin 3
|
|
chr1_+_234372454
|
1.281
|
NM_003272
|
GPR137B
|
G protein-coupled receptor 137B
|
|
chr5_+_17270781
|
1.277
|
|
BASP1
|
brain abundant, membrane attached signal protein 1
|
|
chr16_+_31390567
|
1.259
|
NM_001042454
NM_001164719
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr1_-_27871306
|
1.254
|
NM_002038
NM_022872
NM_022873
|
IFI6
|
interferon, alpha-inducible protein 6
|
|
chr13_-_32678142
|
1.249
|
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr5_+_86600483
|
1.247
|
NM_022650
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1
|
|
chr2_-_38156811
|
1.238
|
NM_000104
|
CYP1B1
|
cytochrome P450, family 1, subfamily B, polypeptide 1
|
|
chr11_+_131285747
|
1.235
|
NM_001144058
NM_001144059
NM_016522
|
NTM
|
neurotrimin
|
|
chr1_-_153209625
|
1.213
|
NM_001130040
NM_183001
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr2_-_38156769
|
1.212
|
|
CYP1B1
|
cytochrome P450, family 1, subfamily B, polypeptide 1
|
|
chr8_-_108579270
|
1.208
|
NM_001146
|
ANGPT1
|
angiopoietin 1
|
|
chr5_-_141973875
|
1.205
|
NM_033137
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr7_+_115953582
|
1.192
|
NM_001172896
NM_001172897
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr12_+_52708460
|
1.187
|
NM_004503
|
HOXC6
|
homeobox C6
|
|
chr10_-_127501683
|
1.149
|
|
UROS
|
uroporphyrinogen III synthase
|
|
chr2_-_1727324
|
1.143
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr10_-_104463947
|
1.140
|
NM_004311
|
ARL3
|
ADP-ribosylation factor-like 3
|
|
chr6_-_80713589
|
1.139
|
NM_022726
|
ELOVL4
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4
|
|
chr3_-_115825742
|
1.131
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr17_-_44037332
|
1.120
|
NM_018952
|
HOXB6
|
homeobox B6
|
|
chr4_-_57671307
|
1.115
|
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chrX_-_13745108
|
1.112
|
|
GPM6B
|
glycoprotein M6B
|
|
chr3_-_55496370
|
1.110
|
NM_003392
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr7_-_81237162
|
1.105
|
NM_000601
NM_001010931
NM_001010932
NM_001010933
NM_001010934
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor)
|
|
chr1_-_153477511
|
1.104
|
|
GBA
|
glucosidase, beta, acid
|
|
chrX_-_10605726
|
1.099
|
NM_001098624
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr11_+_113672407
|
1.098
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr18_-_51219880
|
1.097
|
|
TCF4
|
transcription factor 4
|
|
chr10_+_70745615
|
1.091
|
NM_033496
|
HK1
|
hexokinase 1
|
|
chr4_-_57671273
|
1.084
|
NM_001553
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr11_+_113672378
|
1.078
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr2_-_1727292
|
1.076
|
NM_012293
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr2_-_1727247
|
1.066
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr5_+_119827797
|
1.066
|
|
PRR16
|
proline rich 16
|
|
chr2_-_215967442
|
1.065
|
|
FN1
|
fibronectin 1
|
|
chr20_-_47532585
|
1.064
|
NM_004975
|
KCNB1
|
potassium voltage-gated channel, Shab-related subfamily, member 1
|
|
chr4_-_57671112
|
1.064
|
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr10_+_104493714
|
1.053
|
NM_001083913
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr10_+_70517823
|
1.051
|
NM_002727
|
SRGN
|
serglycin
|
|
chr13_-_37070862
|
1.046
|
NM_001135934
NM_001135935
NM_001135936
NM_006475
|
POSTN
|
periostin, osteoblast specific factor
|
|
chr10_-_127501822
|
1.043
|
NM_000375
|
UROS
|
uroporphyrinogen III synthase
|
|
chrX_-_111953009
|
1.041
|
NM_001113490
|
AMOT
|
angiomotin
|
|
chrX_-_21686079
|
1.036
|
NM_014332
|
SMPX
|
small muscle protein, X-linked
|
|
chr9_+_35819208
|
1.025
|
NM_016446
NM_001042589
|
TMEM8B
|
transmembrane protein 8B
|
|
chr11_+_116575249
|
1.022
|
NM_001001522
NM_003186
|
TAGLN
|
transgelin
|
|
chr11_-_11986704
|
1.020
|
NM_001018057
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr10_+_70517863
|
1.017
|
|
SRGN
|
serglycin
|
|
chr3_-_124822113
|
1.004
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr4_+_169654742
|
0.997
|
NM_001166108
NM_016081
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr13_-_32757835
|
0.995
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr19_+_16296734
|
0.994
|
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr12_+_77782719
|
0.994
|
|
SYT1
|
synaptotagmin I
|
|
chr12_-_90097384
|
0.988
|
NM_133503
|
DCN
|
decorin
|
|
chrX_-_13744933
|
0.983
|
|
GPM6B
|
glycoprotein M6B
|
|
chr10_-_79067210
|
0.962
|
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chr3_-_109423858
|
0.961
|
NM_018010
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas)
|
|
chr4_+_169989680
|
0.960
|
NM_001166110
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr4_+_54790020
|
0.955
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr2_-_237987509
|
0.951
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr3_-_55490401
|
0.950
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr10_+_102782156
|
0.947
|
|
SFXN3
|
sideroflexin 3
|
|
chr1_-_68071635
|
0.944
|
NM_018841
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12
|
|
chr1_+_154095883
|
0.942
|
NM_152280
|
SYT11
|
synaptotagmin XI
|
|
chrX_-_37916138
|
0.938
|
|
SRPX
|
sushi-repeat containing protein, X-linked
|
|
chr2_-_175337386
|
0.936
|
NM_000079
NM_001039523
|
CHRNA1
|
cholinergic receptor, nicotinic, alpha 1 (muscle)
|
|
chr12_-_90096275
|
0.935
|
NM_133504
NM_133505
NM_133506
NM_133507
|
DCN
|
decorin
|
|
chr16_+_83411086
|
0.932
|
NM_031476
|
CRISPLD2
|
cysteine-rich secretory protein LCCL domain containing 2
|
|
chr10_+_3141596
|
0.928
|
|
PFKP
|
phosphofructokinase, platelet
|
|
chr20_+_9236446
|
0.927
|
NM_000933
|
PLCB4
|
phospholipase C, beta 4
|
|
chr9_+_35819375
|
0.926
|
NM_001042590
|
TMEM8B
|
transmembrane protein 8B
|
|
chr2_-_56004366
|
0.922
|
NM_001039348
NM_001039349
|
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1
|
|
chr4_+_72423633
|
0.917
|
NM_003759
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr6_+_33151703
|
0.912
|
NM_002121
|
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1
|
|
chr1_-_153477673
|
0.911
|
NM_001171812
NM_000157
|
GBA
|
glucosidase, beta, acid
|
|
chr21_+_29439768
|
0.907
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr3_-_124821868
|
0.905
|
|
MYLK
|
myosin light chain kinase
|
|
chr6_+_30018287
|
0.904
|
NM_002116
|
HLA-A
|
major histocompatibility complex, class I, A
|
|
chr2_-_237987579
|
0.898
|
NM_004369
NM_057164
NM_057165
NM_057166
NM_057167
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr12_+_13240868
|
0.894
|
NM_001423
|
EMP1
|
epithelial membrane protein 1
|
|
chr15_+_68971835
|
0.893
|
NM_001199017
NM_017691
NM_001199018
|
LRRC49
|
leucine rich repeat containing 49
|
|
chr7_+_90176647
|
0.888
|
NM_012395
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr6_-_112147946
|
0.884
|
NM_153047
|
FYN
|
FYN oncogene related to SRC, FGR, YES
|
|
chr11_+_112337199
|
0.882
|
NM_000615
NM_001076682
NM_181351
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr5_+_119827903
|
0.880
|
NM_016644
|
PRR16
|
proline rich 16
|
|
chr11_-_8849485
|
0.876
|
|
ST5
|
suppression of tumorigenicity 5
|
|
chr5_+_125723686
|
0.873
|
NM_001146319
|
GRAMD3
|
GRAM domain containing 3
|
|
chr1_-_42738535
|
0.870
|
|
TMSB4XP1
|
thymosin beta 4, X-linked pseudogene 1
|
|
chr9_+_113698844
|
0.863
|
NM_003358
|
UGCG
|
UDP-glucose ceramide glucosyltransferase
|
|
chr10_-_92671005
|
0.862
|
NM_014391
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chrX_+_51503219
|
0.850
|
NM_018094
|
GSPT2
|
G1 to S phase transition 2
|
|
chr12_+_46799279
|
0.848
|
NM_000289
|
PFKM
|
phosphofructokinase, muscle
|
|
chr8_-_119192985
|
0.843
|
NM_000127
|
EXT1
|
exostosin 1
|
|
chr10_+_102782151
|
0.840
|
|
SFXN3
|
sideroflexin 3
|
|
chr15_-_81667169
|
0.838
|
NM_016073
|
HDGFRP3
|
hepatoma-derived growth factor, related protein 3
|
|
chr10_-_105202139
|
0.838
|
NM_015916
|
CALHM2
|
calcium homeostasis modulator 2
|
|
chr5_+_140733664
|
0.837
|
NM_018919
NM_032086
|
PCDHGA6
|
protocadherin gamma subfamily A, 6
|
|
chr8_+_37773915
|
0.834
|
|
GPR124
|
G protein-coupled receptor 124
|
|
chr3_-_115960807
|
0.829
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr11_-_14337167
|
0.819
|
NM_012250
|
RRAS2
|
related RAS viral (r-ras) oncogene homolog 2
|
|
chr11_-_64403712
|
0.818
|
|
EHD1
|
EH-domain containing 1
|
|
chr8_-_93099040
|
0.809
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr7_+_98084531
|
0.799
|
NM_002523
|
NPTX2
|
neuronal pentraxin II
|
|
chr5_+_131621230
|
0.799
|
NM_001131027
NM_003687
|
PDLIM4
|
PDZ and LIM domain 4
|
|
chr10_-_79067400
|
0.798
|
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chr5_-_172688024
|
0.795
|
|
STC2
|
stanniocalcin 2
|
|
chr5_+_36644187
|
0.794
|
NM_001166695
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr4_+_84175262
|
0.781
|
NM_016129
|
COPS4
|
COP9 constitutive photomorphogenic homolog subunit 4 (Arabidopsis)
|
|
chr2_-_237987554
|
0.775
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chrX_-_106130098
|
0.773
|
NM_001085354
NM_024657
|
MORC4
|
MORC family CW-type zinc finger 4
|
|
chr5_+_53787187
|
0.772
|
NM_006308
|
HSPB3
|
heat shock 27kDa protein 3
|
|
chrX_+_135106578
|
0.770
|
NM_001159701
NM_001159699
|
FHL1
|
four and a half LIM domains 1
|
|
chr7_-_139948780
|
0.770
|
NM_015689
|
DENND2A
|
DENN/MADD domain containing 2A
|
|
chr5_+_131621276
|
0.770
|
|
PDLIM4
|
PDZ and LIM domain 4
|
|
chr5_-_81082699
|
0.769
|
NM_012446
|
SSBP2
|
single-stranded DNA binding protein 2
|
|
chr3_-_45242072
|
0.769
|
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chr13_+_109932934
|
0.763
|
|
COL4A2
|
collagen, type IV, alpha 2
|