|
chr15_+_37660568
|
2.399
|
NM_003246
|
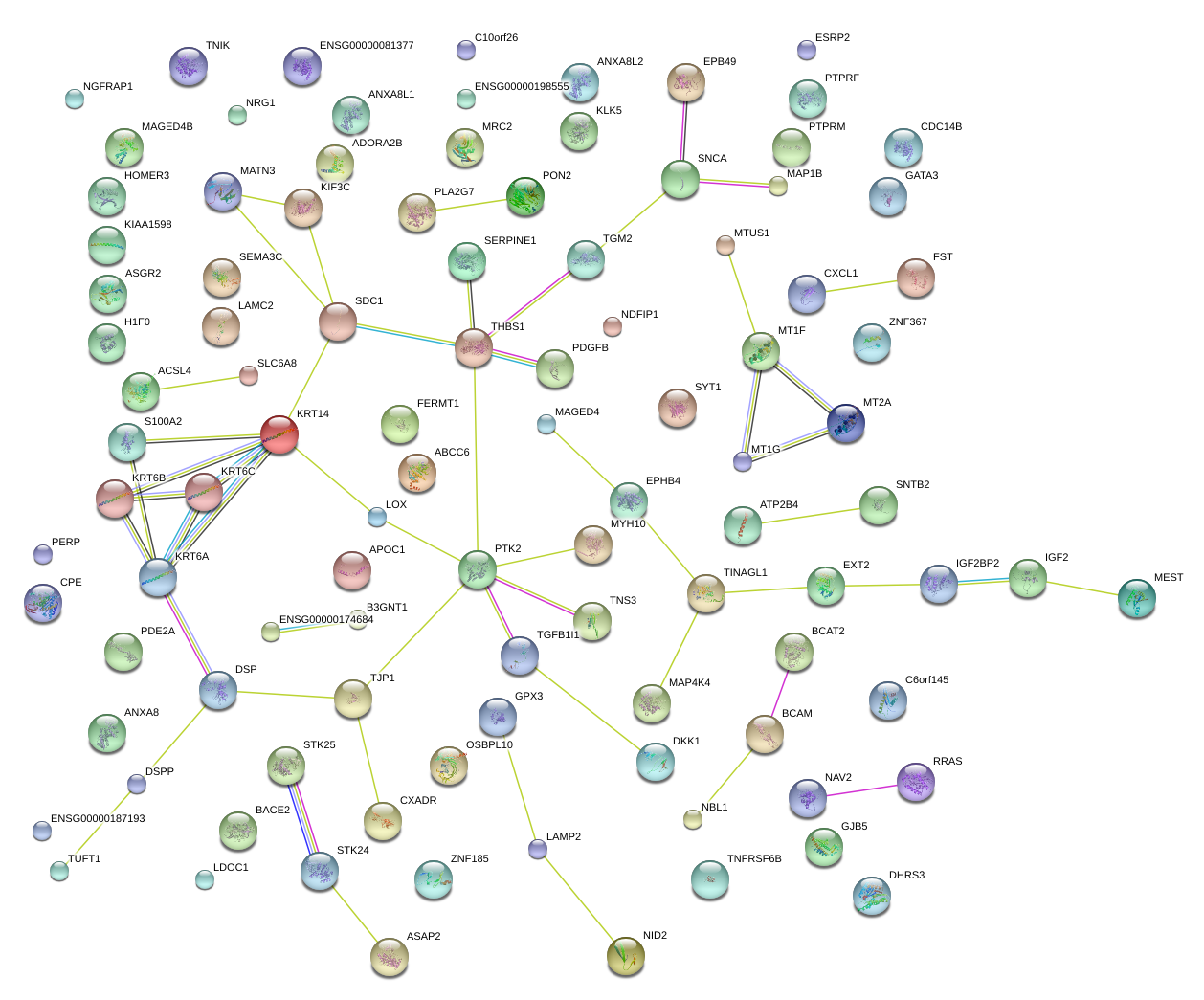
THBS1
|
thrombospondin 1
|
|
chr3_-_31998241
|
1.655
|
NM_001174060
NM_017784
|
OSBPL10
|
oxysterol binding protein-like 10
|
|
chrX_+_55951280
|
1.490
|
|
|
|
|
chr10_+_53744043
|
1.303
|
NM_012242
|
DKK1
|
dickkopf homolog 1 (Xenopus laevis)
|
|
chr19_-_54835162
|
1.302
|
NM_006270
|
RRAS
|
related RAS viral (r-ras) oncogene homolog
|
|
chr7_+_100557098
|
1.288
|
NM_000602
NM_001165413
|
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1
|
|
chr6_-_3697244
|
1.287
|
NM_183373
|
C6orf145
|
chromosome 6 open reading frame 145
|
|
chr1_+_43837096
|
1.196
|
|
PTPRF
|
protein tyrosine phosphatase, receptor type, F
|
|
chr17_+_15788955
|
1.145
|
NM_000676
|
ADORA2B
|
adenosine A2b receptor
|
|
chr2_+_9264360
|
1.141
|
|
ASAP2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2
|
|
chr3_-_31997738
|
1.138
|
|
OSBPL10
|
oxysterol binding protein-like 10
|
|
chr3_-_187025501
|
1.081
|
NM_001007225
NM_006548
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr5_-_121440704
|
1.067
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr16_+_31390567
|
1.050
|
NM_001042454
NM_001164719
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr8_-_17599438
|
1.021
|
NM_020749
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr11_+_44074322
|
1.010
|
NM_000401
|
EXT2
|
exostosin 2
|
|
chr2_+_9264327
|
0.990
|
NM_001135191
NM_003887
|
ASAP2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2
|
|
chr5_+_52812165
|
0.985
|
|
FST
|
follistatin
|
|
chr21_+_17807167
|
0.948
|
NM_001338
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr10_-_46593923
|
0.945
|
NM_001040084
NM_001098845
|
ANXA8L2
LOC653110
ANXA8
ANXA8L1
|
annexin A8-like 2
hypothetical LOC653110
annexin A8
annexin A8-like 1
|
|
chr22_+_36531246
|
0.937
|
|
H1F0
|
H1 histone family, member 0
|
|
chr1_-_151804929
|
0.928
|
NM_005978
|
S100A2
|
S100 calcium binding protein A2
|
|
chr10_+_47216925
|
0.924
|
NM_001630
|
ANXA8L2
ANXA8
|
annexin A8-like 2
annexin A8
|
|
chr20_-_36191623
|
0.877
|
|
TGM2
|
transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase)
|
|
chr8_+_21972631
|
0.848
|
NM_001114138
NM_001978
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr2_-_26058831
|
0.845
|
|
KIF3C
|
kinesin family member 3C
|
|
chr8_+_21972804
|
0.833
|
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr14_-_51605695
|
0.826
|
NM_007361
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr6_+_7486836
|
0.793
|
|
DSP
|
desmoplakin
|
|
chr2_-_20075889
|
0.788
|
|
MATN3
|
matrilin 3
|
|
chr9_-_98368913
|
0.787
|
NM_001077181
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr1_+_201862552
|
0.786
|
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4
|
|
chr5_+_71438796
|
0.785
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr4_+_166519392
|
0.780
|
NM_001873
|
CPE
|
carboxypeptidase E
|
|
chr20_-_6052056
|
0.779
|
NM_017671
|
FERMT1
|
fermitin family member 1
|
|
chr6_+_7486891
|
0.771
|
|
DSP
|
desmoplakin
|
|
chr8_+_31616809
|
0.760
|
NM_013962
|
NRG1
|
neuregulin 1
|
|
chr14_-_51605487
|
0.745
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr22_+_36531178
|
0.742
|
|
H1F0
|
H1 histone family, member 0
|
|
chr11_+_19691397
|
0.741
|
NM_145117
NM_182964
|
NAV2
|
neuron navigator 2
|
|
chrX_+_152608159
|
0.734
|
NM_001142806
|
SLC6A8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8
|
|
chr5_+_150380325
|
0.725
|
|
GPX3
|
glutathione peroxidase 3 (plasma)
|
|
chr12_-_51132129
|
0.718
|
NM_005555
|
KRT6B
|
keratin 6B
|
|
chr14_-_51605515
|
0.711
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr19_-_18910790
|
0.698
|
NM_001145724
|
HOMER3
|
homer homolog 3 (Drosophila)
|
|
chr7_-_47545717
|
0.692
|
NM_022748
|
TNS3
|
tensin 3
|
|
chr19_+_50109903
|
0.690
|
|
APOC1
|
apolipoprotein C-I
|
|
chr10_+_47875229
|
0.690
|
NM_001040084
NM_001098845
|
ANXA8L2
ANXA8
ANXA8L1
|
annexin A8-like 2
annexin A8
annexin A8-like 1
|
|
chr7_-_80386602
|
0.686
|
NM_006379
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C
|
|
chrX_-_140098941
|
0.676
|
|
LDOC1
|
leucine zipper, down-regulated in cancer 1
|
|
chr5_+_52812236
|
0.672
|
NM_006350
NM_013409
|
FST
|
follistatin
|
|
chr4_-_90977149
|
0.655
|
NM_001146054
NM_007308
NM_000345
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chr2_-_242096673
|
0.644
|
NM_006374
|
STK25
|
serine/threonine kinase 25
|
|
chr14_-_51605461
|
0.640
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr1_+_201862548
|
0.633
|
NM_001001396
NM_001684
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4
|
|
chrX_-_140098959
|
0.630
|
NM_012317
|
LDOC1
|
leucine zipper, down-regulated in cancer 1
|
|
chr1_-_12599934
|
0.629
|
NM_004753
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3
|
|
chr1_+_19842309
|
0.609
|
NM_182744
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr19_-_56147971
|
0.607
|
NM_001077491
NM_001077492
NM_012427
|
KLK5
|
kallikrein-related peptidase 5
|
|
chr1_+_181422016
|
0.606
|
|
LAMC2
|
laminin, gamma 2
|
|
chr11_-_2108035
|
0.598
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr11_-_65871592
|
0.595
|
NM_006876
|
B3GNT1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1
|
|
chr10_-_118754549
|
0.587
|
|
KIAA1598
|
KIAA1598
|
|
chr20_+_61798446
|
0.581
|
NM_003823
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy
|
|
chr2_-_242096122
|
0.580
|
|
STK25
|
serine/threonine kinase 25
|
|
chr7_-_80386258
|
0.576
|
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C
|
|
chrX_-_140099014
|
0.570
|
|
LDOC1
|
leucine zipper, down-regulated in cancer 1
|
|
chr17_-_6958695
|
0.557
|
|
ASGR2
|
asialoglycoprotein receptor 2
|
|
chr17_-_8474733
|
0.552
|
NM_005964
|
MYH10
|
myosin, heavy chain 10, non-muscle
|
|
chr1_-_20682541
|
0.546
|
|
|
|
|
chr3_-_187025438
|
0.546
|
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chrX_+_51944658
|
0.545
|
NM_001098800
NM_030801
NM_177535
NM_177537
|
MAGED4
MAGED4B
|
melanoma antigen family D, 4
melanoma antigen family D, 4B
|
|
chr17_-_36996611
|
0.543
|
NM_000526
|
KRT14
|
keratin 14
|
|
chr7_+_129918408
|
0.540
|
NM_177525
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr16_+_67778441
|
0.536
|
NM_006750
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2)
|
|
chr7_-_94902149
|
0.530
|
NM_000305
NM_001018161
|
PON2
|
paraoxonase 2
|
|
chr17_-_6958773
|
0.526
|
NM_001181
NM_080912
NM_080913
|
ASGR2
|
asialoglycoprotein receptor 2
|
|
chrX_-_119487164
|
0.521
|
|
LAMP2
|
lysosomal-associated membrane protein 2
|
|
chr12_+_77782648
|
0.518
|
|
SYT1
|
synaptotagmin I
|
|
chr16_-_66827449
|
0.516
|
NM_024939
|
ESRP2
|
epithelial splicing regulatory protein 2
|
|
chr16_+_55249355
|
0.515
|
NM_005949
|
MT1F
|
metallothionein 1F
|
|
chrX_-_119487096
|
0.515
|
|
LAMP2
|
lysosomal-associated membrane protein 2
|
|
chr2_+_101874765
|
0.508
|
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr7_-_100262971
|
0.507
|
NM_004444
|
EPHB4
|
EPH receptor B4
|
|
chr5_+_141468568
|
0.505
|
|
NDFIP1
|
Nedd4 family interacting protein 1
|
|
chr2_-_20075929
|
0.503
|
NM_002381
|
MATN3
|
matrilin 3
|
|
chr2_-_242096592
|
0.502
|
|
STK25
|
serine/threonine kinase 25
|
|
chr20_+_61798512
|
0.498
|
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy
|
|
chr10_+_8137356
|
0.495
|
|
GATA3
|
GATA binding protein 3
|
|
chr10_+_8136778
|
0.490
|
|
GATA3
|
GATA binding protein 3
|
|
chr2_-_20288108
|
0.484
|
NM_002997
|
SDC1
|
syndecan 1
|
|
chr6_-_138470160
|
0.483
|
NM_022121
|
PERP
|
PERP, TP53 apoptosis effector
|
|
chr15_-_27901828
|
0.481
|
NM_003257
NM_175610
|
TJP1
|
tight junction protein 1 (zona occludens 1)
|
|
chr7_+_129919157
|
0.477
|
NM_002402
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chrX_+_151837064
|
0.476
|
NM_001178113
|
ZNF185
|
zinc finger protein 185 (LIM domain)
|
|
chr4_+_74953971
|
0.473
|
NM_001511
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha)
|
|
chr8_-_142080169
|
0.471
|
NM_005607
NM_153831
|
PTK2
|
PTK2 protein tyrosine kinase 2
|
|
chr17_-_6958282
|
0.470
|
NM_080914
|
ASGR2
|
asialoglycoprotein receptor 2
|
|
chr10_+_104493714
|
0.469
|
NM_001083913
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr21_+_41461962
|
0.468
|
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chr11_-_72057755
|
0.466
|
NM_001146209
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated
|
|
chr1_+_31814672
|
0.463
|
NM_022164
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chrX_-_108863109
|
0.462
|
|
ACSL4
|
acyl-CoA synthetase long-chain family member 4
|
|
chr1_+_34993234
|
0.461
|
NM_005268
|
GJB5
|
gap junction protein, beta 5, 31.1kDa
|
|
chr17_+_15789147
|
0.460
|
|
ADORA2B
|
adenosine A2b receptor
|
|
chr16_-_16224791
|
0.459
|
NM_001079528
NM_001171
|
ABCC6
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6
|
|
chr18_+_7557313
|
0.446
|
NM_001105244
NM_002845
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chr22_-_37969897
|
0.445
|
|
PDGFB
|
platelet-derived growth factor beta polypeptide (simian sarcoma viral (v-sis) oncogene homolog)
|
|
chrX_-_108863165
|
0.444
|
NM_004458
NM_022977
|
ACSL4
|
acyl-CoA synthetase long-chain family member 4
|
|
chr6_-_46811388
|
0.443
|
NM_001168357
|
PLA2G7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma)
|
|
chr19_+_50109416
|
0.442
|
|
APOC1
|
apolipoprotein C-I
|
|
chrX_+_102518764
|
0.440
|
NM_014380
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr1_+_149779384
|
0.435
|
NM_001126337
NM_020127
|
TUFT1
|
tuftelin 1
|
|
chr17_+_58058783
|
0.431
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr2_+_101874809
|
0.428
|
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr19_+_50004134
|
0.428
|
NM_001013257
NM_005581
|
BCAM
|
basal cell adhesion molecule (Lutheran blood group)
|
|
chr6_+_32021687
|
0.424
|
NM_001710
|
CFB
|
complement factor B
|
|
chr20_+_43468617
|
0.421
|
NM_001048223
NM_001048224
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr12_+_6923441
|
0.421
|
NM_138425
|
C12orf57
|
chromosome 12 open reading frame 57
|
|
chr2_-_20288647
|
0.420
|
NM_001006946
|
SDC1
|
syndecan 1
|
|
chr13_+_112808102
|
0.415
|
NM_000131
NM_019616
|
F7
|
coagulation factor VII (serum prothrombin conversion accelerator)
|
|
chr5_+_71538456
|
0.412
|
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr1_-_20685314
|
0.410
|
NM_018584
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr1_+_31814709
|
0.410
|
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr20_-_22512893
|
0.410
|
|
FOXA2
|
forkhead box A2
|
|
chr7_+_94130673
|
0.408
|
|
PEG10
|
paternally expressed 10
|
|
chr17_-_37022465
|
0.406
|
NM_005557
|
KRT16
|
keratin 16
|
|
chr9_-_112840064
|
0.403
|
NM_001401
NM_057159
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr3_-_130761967
|
0.402
|
|
PLXND1
|
plexin D1
|
|
chr13_+_109935250
|
0.402
|
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr7_-_107883929
|
0.399
|
NM_001193582
NM_001193583
NM_001193584
NM_005010
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr5_+_92946348
|
0.399
|
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr9_+_123101926
|
0.396
|
|
GSN
|
gelsolin
|
|
chr12_+_77782719
|
0.395
|
|
SYT1
|
synaptotagmin I
|
|
chr6_+_7486862
|
0.394
|
NM_001008844
NM_004415
|
DSP
|
desmoplakin
|
|
chr18_+_54013575
|
0.394
|
NM_001144970
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr17_-_74432800
|
0.392
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr4_-_186934059
|
0.388
|
NM_001145673
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr1_-_43005312
|
0.388
|
NM_001146289
NM_022356
|
LEPRE1
|
leucine proline-enriched proteoglycan (leprecan) 1
|
|
chr10_+_120853548
|
0.387
|
|
FAM45A
|
family with sequence similarity 45, member A
|
|
chr2_-_220116508
|
0.384
|
NM_024536
|
CHPF
|
chondroitin polymerizing factor
|
|
chr12_-_45052286
|
0.382
|
|
SLC38A2
|
solute carrier family 38, member 2
|
|
chr2_+_24126140
|
0.382
|
|
FKBP1B
|
FK506 binding protein 1B, 12.6 kDa
|
|
chr2_-_227372574
|
0.382
|
|
IRS1
|
insulin receptor substrate 1
|
|
chr7_+_116099648
|
0.381
|
NM_000245
NM_001127500
|
MET
|
met proto-oncogene (hepatocyte growth factor receptor)
|
|
chr5_-_168204430
|
0.376
|
|
|
|
|
chr5_+_176449156
|
0.375
|
NM_022963
|
FGFR4
|
fibroblast growth factor receptor 4
|
|
chr15_+_88610181
|
0.374
|
|
NGRN
|
neugrin, neurite outgrowth associated
|
|
chr6_+_83130220
|
0.373
|
|
TPBG
|
trophoblast glycoprotein
|
|
chr3_+_187917791
|
0.373
|
NM_000893
NM_001102416
NM_001166451
|
KNG1
|
kininogen 1
|
|
chr1_+_20788027
|
0.372
|
NM_001785
|
CDA
|
cytidine deaminase
|
|
chr16_-_62573
|
0.372
|
NM_022450
|
RHBDF1
|
rhomboid 5 homolog 1 (Drosophila)
|
|
chr2_+_24126087
|
0.372
|
NM_004116
NM_054033
|
FKBP1B
|
FK506 binding protein 1B, 12.6 kDa
|
|
chr13_+_113574925
|
0.368
|
NM_001143945
|
GAS6
|
growth arrest-specific 6
|
|
chr17_-_77634455
|
0.367
|
|
FASN
|
fatty acid synthase
|
|
chrX_-_119487183
|
0.364
|
NM_001122606
NM_002294
NM_013995
|
LAMP2
|
lysosomal-associated membrane protein 2
|
|
chr21_-_43903755
|
0.363
|
NM_007031
|
HSF2BP
|
heat shock transcription factor 2 binding protein
|
|
chr12_-_51200347
|
0.362
|
NM_000424
|
KRT5
|
keratin 5
|
|
chr12_-_45052830
|
0.361
|
NM_018976
|
SLC38A2
|
solute carrier family 38, member 2
|
|
chr1_-_180621272
|
0.359
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr1_+_31814712
|
0.359
|
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr22_-_36998889
|
0.359
|
|
TMEM184B
|
transmembrane protein 184B
|
|
chr22_-_17891777
|
0.356
|
|
CLDN5
|
claudin 5
|
|
chr4_+_1764802
|
0.355
|
NM_000142
NM_001163213
NM_022965
|
FGFR3
|
fibroblast growth factor receptor 3
|
|
chr18_+_19706979
|
0.354
|
NM_000227
NM_001127718
|
LAMA3
|
laminin, alpha 3
|
|
chr14_-_34169047
|
0.353
|
NM_021249
NM_152233
|
SNX6
|
sorting nexin 6
|
|
chr10_+_5556923
|
0.353
|
NM_005185
|
CALML3
|
calmodulin-like 3
|
|
chr11_+_46359781
|
0.349
|
NM_002391
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr13_+_112789669
|
0.348
|
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr16_+_27232745
|
0.348
|
NM_000418
NM_001008699
|
IL4R
|
interleukin 4 receptor
|
|
chr3_-_10042989
|
0.347
|
|
CIDECP
|
cell death-inducing DFFA-like effector c pseudogene
|
|
chr1_+_36462583
|
0.346
|
NM_005119
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr20_+_3399615
|
0.346
|
NM_139321
NM_139322
|
ATRN
|
attractin
|
|
chr1_-_55125469
|
0.345
|
|
DHCR24
|
24-dehydrocholesterol reductase
|
|
chr2_-_220144427
|
0.344
|
|
OBSL1
|
obscurin-like 1
|
|
chr22_-_36998912
|
0.343
|
NM_001195072
NM_012264
|
TMEM184B
|
transmembrane protein 184B
|
|
chr17_+_37372222
|
0.343
|
NM_033133
|
CNP
|
2',3'-cyclic nucleotide 3' phosphodiesterase
|
|
chr2_-_175578156
|
0.340
|
NM_001025201
NM_001822
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr9_+_123101701
|
0.339
|
NM_000177
|
GSN
|
gelsolin
|
|
chr14_-_104515738
|
0.339
|
NM_138420
|
AHNAK2
|
AHNAK nucleoprotein 2
|
|
chr5_-_175776137
|
0.338
|
NM_001834
NM_007097
|
CLTB
|
clathrin, light chain B
|
|
chrX_+_43400350
|
0.335
|
NM_000240
|
MAOA
|
monoamine oxidase A
|
|
chr2_+_188865796
|
0.334
|
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1
|
|
chr6_-_2785682
|
0.334
|
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1
|
|
chr2_+_188865634
|
0.333
|
NM_016315
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1
|
|
chr11_+_46359836
|
0.333
|
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr2_+_10361276
|
0.331
|
NM_134421
|
HPCAL1
|
hippocalcin-like 1
|
|
chr9_+_35663852
|
0.328
|
NM_001216
|
CA9
|
carbonic anhydrase IX
|
|
chr8_-_134653194
|
0.327
|
NM_003033
NM_173344
|
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1
|
|
chr10_+_121401061
|
0.324
|
|
BAG3
|
BCL2-associated athanogene 3
|
|
chr2_+_241023703
|
0.322
|
NM_002081
|
GPC1
|
glypican 1
|
|
chr17_-_1342700
|
0.321
|
NM_001080779
|
MYO1C
|
myosin IC
|
|
chr2_-_26058941
|
0.319
|
NM_002254
|
KIF3C
|
kinesin family member 3C
|
|
chr16_+_56259657
|
0.319
|
NM_170776
|
GPR97
|
G protein-coupled receptor 97
|
|
chr16_+_54072969
|
0.319
|
NM_001127891
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr1_-_43005243
|
0.318
|
|
LEPRE1
|
leucine proline-enriched proteoglycan (leprecan) 1
|
|
chr12_-_51173238
|
0.318
|
NM_005554
|
KRT6A
|
keratin 6A
|
|
chr1_+_218930542
|
0.317
|
|
C1orf115
|
chromosome 1 open reading frame 115
|
|
chr21_+_41461597
|
0.316
|
NM_012105
NM_138991
NM_138992
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chr6_-_166960690
|
0.315
|
NM_021135
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|