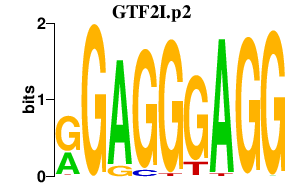
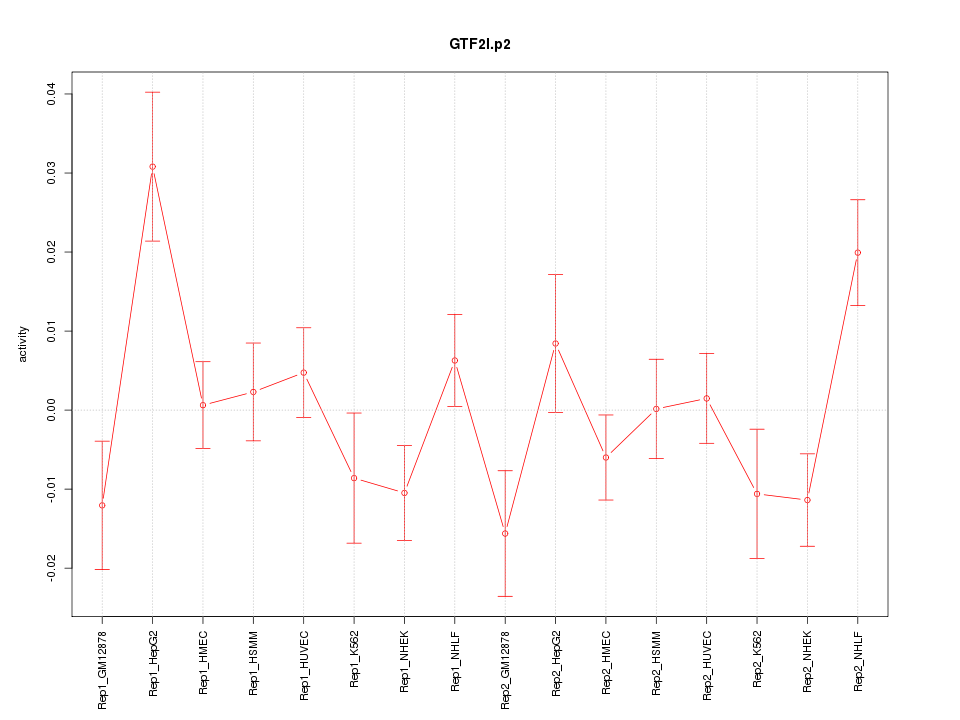
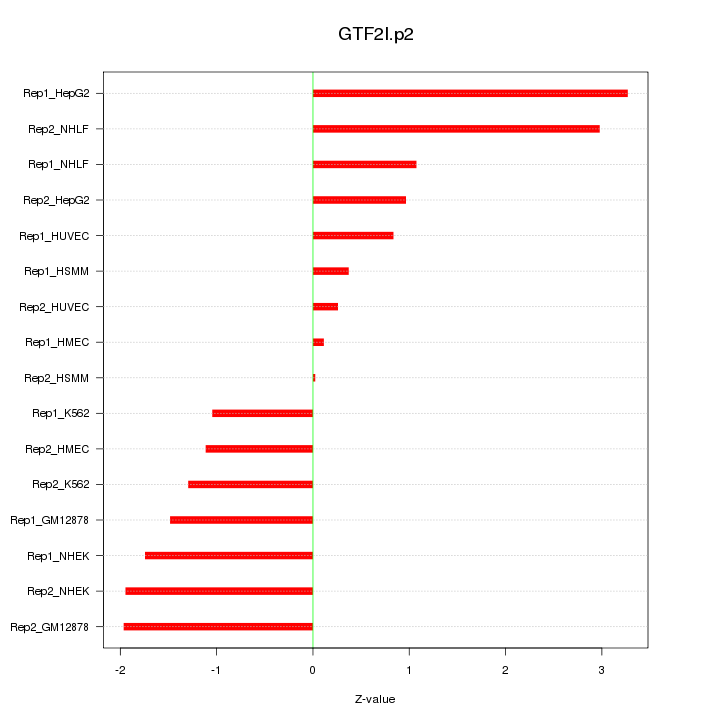
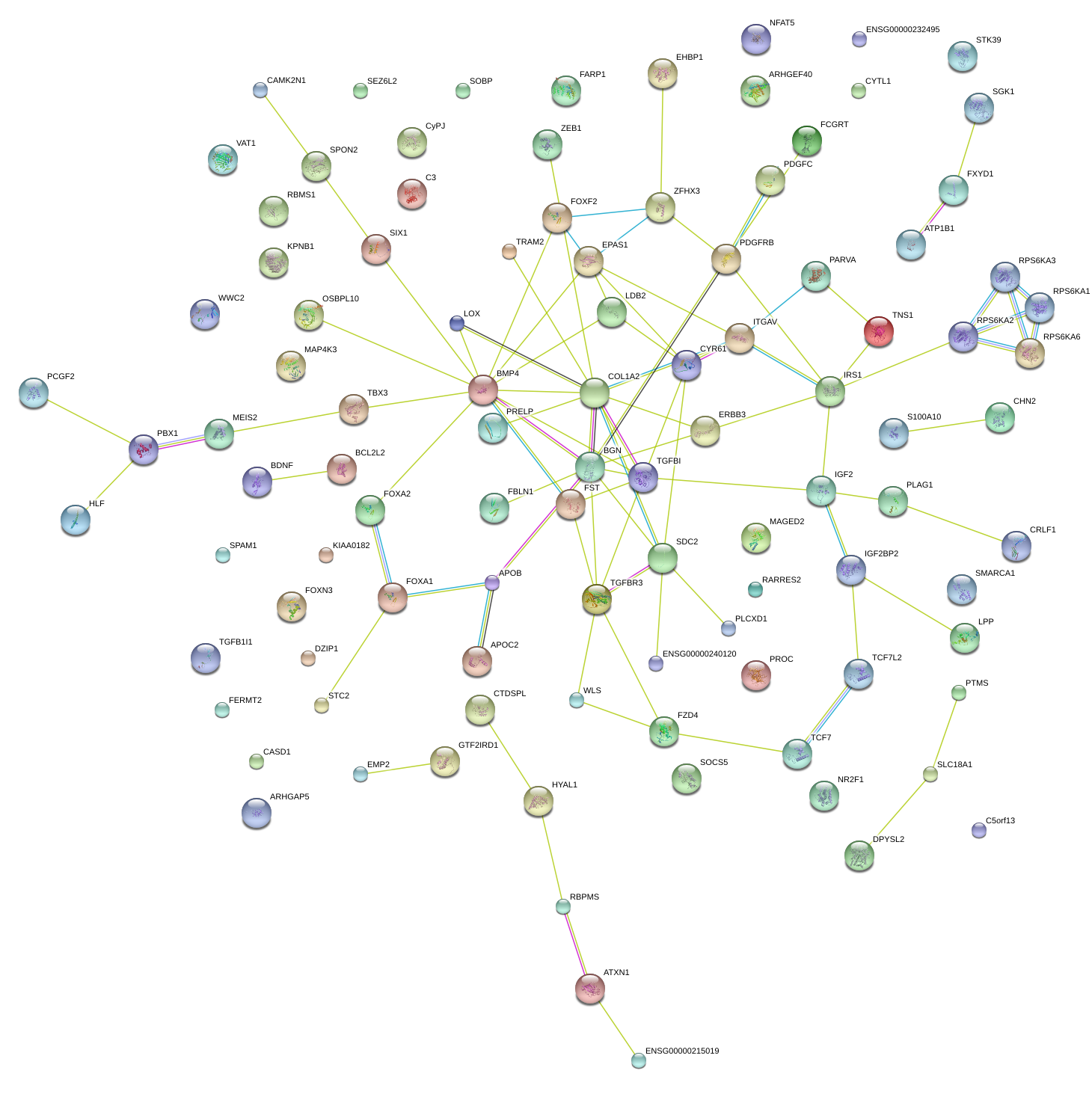
|
chr1_-_20685314
|
5.211
|
NM_018584
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr2_-_227371698
|
3.435
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr2_-_218551786
|
3.051
|
|
TNS1
|
tensin 1
|
|
chr5_+_135392482
|
2.936
|
NM_000358
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa
|
|
chr8_+_97575317
|
2.781
|
|
SDC2
|
syndecan 2
|
|
chr8_+_97575196
|
2.768
|
|
SDC2
|
syndecan 2
|
|
chr8_+_97575244
|
2.764
|
|
SDC2
|
syndecan 2
|
|
chr2_-_39517934
|
2.540
|
|
MAP4K3
|
mitogen-activated protein kinase kinase kinase kinase 3
|
|
chr19_+_50144267
|
2.510
|
|
APOC2
|
apolipoprotein C-II
|
|
chr12_-_113606322
|
2.435
|
NM_005996
NM_016569
|
TBX3
|
T-box 3
|
|
chr2_-_39517795
|
2.413
|
|
MAP4K3
|
mitogen-activated protein kinase kinase kinase kinase 3
|
|
chr3_-_187025501
|
2.296
|
NM_001007225
NM_006548
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr10_+_114699953
|
2.234
|
NM_001146274
NM_001146283
NM_001146284
NM_001146285
NM_001146286
NM_001198525
NM_001198526
NM_001198527
NM_001198528
NM_001198529
NM_001198530
NM_001198531
NM_030756
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr5_+_92944680
|
2.214
|
NM_005654
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr14_-_53493198
|
2.060
|
NM_001202
NM_130850
|
BMP4
|
bone morphogenetic protein 4
|
|
chr2_-_39517633
|
2.022
|
NM_003618
|
MAP4K3
|
mitogen-activated protein kinase kinase kinase kinase 3
|
|
chr22_-_34566276
|
2.011
|
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chrX_-_128485134
|
1.937
|
NM_003069
NM_139035
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chr4_-_1156959
|
1.915
|
NM_001128325
|
SPON2
|
spondin 2, extracellular matrix protein
|
|
chr22_-_34566210
|
1.892
|
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr11_-_2116048
|
1.869
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr8_+_97575370
|
1.866
|
|
SDC2
|
syndecan 2
|
|
chrX_+_54851500
|
1.842
|
NM_014599
|
MAGED2
|
melanoma antigen family D, 2
|
|
chr20_-_22512893
|
1.790
|
|
FOXA2
|
forkhead box A2
|
|
chrX_+_138060
|
1.769
|
NM_018390
|
PLCXD1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1
|
|
chrY_+_138060
|
1.769
|
NM_018390
|
PLCXD1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1
|
|
chr8_+_97575052
|
1.741
|
NM_002998
|
SDC2
|
syndecan 2
|
|
chr2_-_227372574
|
1.680
|
|
IRS1
|
insulin receptor substrate 1
|
|
chr2_-_168812132
|
1.673
|
NM_013233
|
STK39
|
serine threonine kinase 39
|
|
chr5_-_111120817
|
1.636
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chrX_-_128485121
|
1.622
|
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chr11_-_2116775
|
1.608
|
NM_000612
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr1_-_92124201
|
1.595
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr3_+_189354167
|
1.588
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr16_-_29817841
|
1.566
|
NM_001114099
NM_001114100
NM_012410
NM_201575
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2
|
|
chr8_+_26427378
|
1.556
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr22_-_34566367
|
1.547
|
NM_001031695
NM_001082576
NM_001082577
NM_014309
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr4_-_16509446
|
1.544
|
NM_001130834
NM_001290
|
LDB2
|
LIM domain binding 2
|
|
chr4_-_16509354
|
1.518
|
|
LDB2
|
LIM domain binding 2
|
|
chr11_-_2116501
|
1.511
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr4_-_1156533
|
1.507
|
NM_012445
|
SPON2
|
spondin 2, extracellular matrix protein
|
|
chrX_-_128485062
|
1.492
|
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chr7_-_149669622
|
1.463
|
NM_002889
|
RARRES2
|
retinoic acid receptor responder (tazarotene induced) 2
|
|
chr2_+_62786290
|
1.460
|
NM_001142616
NM_015252
|
EHBP1
|
EH domain binding protein 1
|
|
chr11_-_2116360
|
1.456
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr5_-_111120929
|
1.456
|
NM_001142482
NM_001142481
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr2_+_187162969
|
1.455
|
NM_001145000
NM_002210
|
ITGAV
|
integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51)
|
|
chr14_+_20608179
|
1.454
|
NM_018071
|
ARHGEF40
|
Rho guanine nucleotide exchange factor (GEF) 40
|
|
chr1_-_92124064
|
1.438
|
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr2_+_127892459
|
1.432
|
NM_000312
|
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa)
|
|
chr13_+_97593469
|
1.418
|
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr3_-_187025438
|
1.410
|
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr1_+_85819004
|
1.406
|
NM_001554
|
CYR61
|
cysteine-rich, angiogenic inducer, 61
|
|
chr5_-_111120764
|
1.402
|
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr2_+_127892489
|
1.397
|
|
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa)
|
|
chr13_+_97593411
|
1.394
|
NM_001001715
NM_005766
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr6_+_1335067
|
1.378
|
NM_001452
|
FOXF2
|
forkhead box F2
|
|
chr14_+_31616196
|
1.369
|
NM_001030055
NM_001173
|
ARHGAP5
|
Rho GTPase activating protein 5
|
|
chr11_-_27697768
|
1.357
|
NM_001143807
|
BDNF
|
brain-derived neurotrophic factor
|
|
chrX_-_20194831
|
1.314
|
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3
|
|
chr5_-_172689111
|
1.280
|
NM_003714
|
STC2
|
stanniocalcin 2
|
|
chr17_+_50697355
|
1.274
|
|
HLF
|
hepatic leukemia factor
|
|
chr19_+_40321567
|
1.268
|
NM_021902
|
FXYD1
|
FXYD domain containing ion transport regulator 1
|
|
chr19_+_40321584
|
1.266
|
|
FXYD1
|
FXYD domain containing ion transport regulator 1
|
|
chr16_+_68157370
|
1.258
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr13_+_97593711
|
1.250
|
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr2_+_46779827
|
1.244
|
NM_144949
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr1_-_150233337
|
1.234
|
NM_002966
|
S100A10
|
S100 calcium binding protein A10
|
|
chr17_-_34157962
|
1.225
|
NM_007144
|
PCGF2
|
polycomb group ring finger 2
|
|
chr4_-_81213207
|
1.221
|
|
|
|
|
chr3_-_31997738
|
1.205
|
|
OSBPL10
|
oxysterol binding protein-like 10
|
|
chr16_-_10581944
|
1.202
|
NM_001424
|
EMP2
|
epithelial membrane protein 2
|
|
chr12_+_54760153
|
1.199
|
NM_001005915
NM_001982
|
ERBB3
|
v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian)
|
|
chr19_+_54708247
|
1.198
|
NM_004107
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr1_-_150232900
|
1.179
|
|
S100A10
|
S100 calcium binding protein A10
|
|
chr14_+_22845851
|
1.167
|
NM_004050
|
BCL2L2-PABPN1
BCL2L2
|
BCL2L2-PABPN1 read-through transcript
BCL2-like 2
|
|
chr17_-_38421363
|
1.163
|
|
VAT1
|
vesicle amine transport protein 1 homolog (T. californica)
|
|
chr11_-_86343859
|
1.158
|
NM_012193
|
FZD4
|
frizzled homolog 4 (Drosophila)
|
|
chr8_+_97575071
|
1.150
|
|
SDC2
|
syndecan 2
|
|
chr14_-_53491019
|
1.148
|
NM_130851
|
BMP4
|
bone morphogenetic protein 4
|
|
chr2_-_161058550
|
1.134
|
NM_002897
NM_016836
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr19_+_54708312
|
1.125
|
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr4_+_184257337
|
1.120
|
NM_024949
|
WWC2
|
WW and C2 domain containing 2
|
|
chr10_+_31648103
|
1.108
|
NM_001174093
NM_001174095
NM_001174096
NM_030751
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr13_-_95094920
|
1.106
|
NM_014934
NM_198968
|
DZIP1
|
DAZ interacting protein 1
|
|
chr6_-_16869579
|
1.069
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chrX_+_152413540
|
1.067
|
NM_001711
|
BGN
|
biglycan
|
|
chr22_-_34754343
|
1.053
|
NM_001082578
NM_001082579
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr19_-_6671585
|
1.049
|
NM_000064
|
C3
|
complement component 3
|
|
chr5_+_52812165
|
1.048
|
|
FST
|
follistatin
|
|
chr6_-_134538762
|
1.034
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr1_-_68470585
|
1.024
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chr7_+_29200636
|
0.993
|
NM_004067
|
CHN2
|
chimerin (chimaerin) 2
|
|
chr14_-_52487558
|
0.991
|
NM_001134999
NM_001135000
NM_006832
|
FERMT2
|
fermitin family member 2
|
|
chr19_-_18578577
|
0.974
|
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr16_-_71650034
|
0.970
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr4_-_16509126
|
0.957
|
|
LDB2
|
LIM domain binding 2
|
|
chr5_-_149515331
|
0.955
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr14_-_88953059
|
0.944
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr16_+_84202515
|
0.942
|
NM_001134473
|
KIAA0182
|
KIAA0182
|
|
chr8_+_30361485
|
0.938
|
NM_001008710
NM_001008711
NM_001008712
NM_006867
|
RBPMS
|
RNA binding protein with multiple splicing
|
|
chr7_+_93976927
|
0.931
|
NM_022900
|
CASD1
|
CAS1 domain containing 1
|
|
chr5_-_121441910
|
0.928
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr16_+_68157611
|
0.917
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr5_+_52812236
|
0.912
|
NM_006350
NM_013409
|
FST
|
follistatin
|
|
chr6_-_52549774
|
0.903
|
NM_012288
|
TRAM2
|
translocation associated membrane protein 2
|
|
chr4_-_1156378
|
0.901
|
|
SPON2
|
spondin 2, extracellular matrix protein
|
|
chr6_-_134538526
|
0.895
|
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr1_+_167342493
|
0.893
|
NM_001677
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr1_+_162795328
|
0.889
|
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr4_-_1156362
|
0.888
|
|
SPON2
|
spondin 2, extracellular matrix protein
|
|
chr4_-_158111995
|
0.887
|
NM_016205
|
PDGFC
|
platelet derived growth factor C
|
|
chr3_+_37878647
|
0.872
|
NM_001008392
NM_005808
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like
|
|
chr1_+_167342149
|
0.868
|
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr2_-_21120314
|
0.861
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr15_-_35179614
|
0.861
|
|
MEIS2
|
Meis homeobox 2
|
|
chr11_+_12355600
|
0.861
|
NM_018222
|
PARVA
|
parvin, alpha
|
|
chr1_+_201711578
|
0.860
|
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein
|
|
chr17_-_7095982
|
0.856
|
NM_015343
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1
|
|
chrX_-_20195062
|
0.839
|
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3
|
|
chr22_+_44277354
|
0.831
|
NM_001996
NM_006485
NM_006486
NM_006487
|
FBLN1
|
fibulin 1
|
|
chr17_+_43082302
|
0.831
|
|
KPNB1
|
karyopherin (importin) beta 1
|
|
chr14_-_37134191
|
0.826
|
NM_004496
|
FOXA1
|
forkhead box A1
|
|
chr7_+_93862137
|
0.820
|
|
COL1A2
PLAG1
|
collagen, type I, alpha 2
pleiomorphic adenoma gene 1
|
|
chr14_-_60185659
|
0.820
|
NM_005982
|
SIX1
|
SIX homeobox 1
|
|
chr7_+_73506172
|
0.810
|
|
GTF2IRD1
|
GTF2I repeat domain containing 1
|
|
chr19_+_54707739
|
0.809
|
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr6_+_107917965
|
0.805
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chrX_+_152413646
|
0.804
|
|
BGN
|
biglycan
|
|
chr16_+_31390567
|
0.803
|
NM_001042454
NM_001164719
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr3_-_50324815
|
0.798
|
NM_153281
|
HYAL1
|
hyaluronoglucosaminidase 1
|
|
chr7_+_93977267
|
0.795
|
|
CASD1
|
CAS1 domain containing 1
|
|
chr3_-_8668723
|
0.793
|
NM_015931
|
C3orf32
|
chromosome 3 open reading frame 32
|
|
chr3_+_23961739
|
0.783
|
NM_005126
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr7_-_107430566
|
0.779
|
NM_002291
|
LAMB1
|
laminin, beta 1
|
|
chr7_-_94123362
|
0.778
|
|
SGCE
|
sarcoglycan, epsilon
|
|
chr3_+_54131731
|
0.777
|
NM_018398
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3
|
|
chr8_+_22492198
|
0.777
|
NM_021630
|
PDLIM2
|
PDZ and LIM domain 2 (mystique)
|
|
chr11_-_70350292
|
0.776
|
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr7_+_93861808
|
0.774
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr7_-_94123413
|
0.768
|
NM_001099400
NM_001099401
NM_003919
|
SGCE
|
sarcoglycan, epsilon
|
|
chr17_+_64020155
|
0.764
|
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1)
|
|
chr17_+_38088097
|
0.764
|
NM_003632
|
CNTNAP1
|
contactin associated protein 1
|
|
chr6_-_31805547
|
0.761
|
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2
|
|
chr2_-_174537021
|
0.757
|
NM_001017371
|
SP3
|
Sp3 transcription factor
|
|
chr2_+_46779553
|
0.756
|
NM_014011
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr7_-_94123308
|
0.754
|
|
SGCE
|
sarcoglycan, epsilon
|
|
chr20_-_22513050
|
0.752
|
NM_021784
|
FOXA2
|
forkhead box A2
|
|
chr11_-_2108035
|
0.751
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr16_-_63713382
|
0.747
|
NM_001797
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr19_+_40213431
|
0.747
|
NM_001037
NM_199037
|
SCN1B
|
sodium channel, voltage-gated, type I, beta
|
|
chr12_-_123023312
|
0.738
|
|
CCDC92
|
coiled-coil domain containing 92
|
|
chr2_+_54536780
|
0.736
|
NM_003128
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|
|
chr3_-_126088530
|
0.731
|
|
ITGB5
|
integrin, beta 5
|
|
chr1_+_203740306
|
0.728
|
NM_002596
NM_212502
NM_212503
|
CDK18
|
cyclin-dependent kinase 18
|
|
chr17_+_25730107
|
0.725
|
NM_001304
|
CPD
|
carboxypeptidase D
|
|
chr3_+_54131613
|
0.724
|
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3
|
|
chrX_-_130251068
|
0.722
|
NM_001170961
NM_001170962
NM_001555
NM_205833
|
IGSF1
|
immunoglobulin superfamily, member 1
|
|
chr12_-_105165805
|
0.719
|
NM_006825
|
CKAP4
|
cytoskeleton-associated protein 4
|
|
chr16_-_63713466
|
0.719
|
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr19_-_53586549
|
0.712
|
NM_006801
|
KDELR1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1
|
|
chr1_-_150232927
|
0.708
|
|
S100A10
|
S100 calcium binding protein A10
|
|
chr9_-_16860719
|
0.705
|
NM_017637
|
BNC2
|
basonuclin 2
|
|
chr5_-_149515613
|
0.705
|
NM_002609
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr21_+_39099624
|
0.702
|
|
ETS2
|
v-ets erythroblastosis virus E26 oncogene homolog 2 (avian)
|
|
chr5_+_150380325
|
0.701
|
|
GPX3
|
glutathione peroxidase 3 (plasma)
|
|
chr16_-_63713485
|
0.700
|
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr8_-_134653194
|
0.698
|
NM_003033
NM_173344
|
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1
|
|
chr6_+_151602787
|
0.693
|
NM_005100
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr1_+_201711505
|
0.693
|
NM_002725
NM_201348
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein
|
|
chr1_+_202308814
|
0.691
|
NM_005686
|
SOX13
|
SRY (sex determining region Y)-box 13
|
|
chr2_-_21120420
|
0.690
|
NM_000384
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr15_-_35179650
|
0.688
|
NM_170674
NM_170675
NM_170676
NM_170677
|
MEIS2
|
Meis homeobox 2
|
|
chr2_-_166940710
|
0.684
|
NM_002977
|
SCN9A
|
sodium channel, voltage-gated, type IX, alpha subunit
|
|
chr10_-_62373828
|
0.683
|
NM_014836
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chr20_+_43468617
|
0.679
|
NM_001048223
NM_001048224
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr3_+_106568430
|
0.677
|
|
ALCAM
|
activated leukocyte cell adhesion molecule
|
|
chr6_+_151603201
|
0.669
|
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr19_-_53586427
|
0.667
|
|
KDELR1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1
|
|
chr2_+_36436873
|
0.665
|
NM_016441
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr19_+_54708302
|
0.664
|
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr5_-_149515502
|
0.664
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr12_+_54760211
|
0.664
|
|
ERBB3
|
v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian)
|
|
chr12_-_45052286
|
0.660
|
|
SLC38A2
|
solute carrier family 38, member 2
|
|
chr1_+_212228482
|
0.655
|
NM_002763
|
PROX1
|
prospero homeobox 1
|
|
chr17_+_50697319
|
0.650
|
NM_002126
|
HLF
|
hepatic leukemia factor
|
|
chr7_+_73506047
|
0.646
|
NM_016328
|
GTF2IRD1
|
GTF2I repeat domain containing 1
|
|
chr15_-_59308708
|
0.645
|
|
RORA
|
RAR-related orphan receptor A
|
|
chr17_+_43082273
|
0.644
|
NM_002265
|
KPNB1
|
karyopherin (importin) beta 1
|
|
chr21_+_39099717
|
0.643
|
NM_005239
|
ETS2
|
v-ets erythroblastosis virus E26 oncogene homolog 2 (avian)
|
|
chr7_+_100557098
|
0.640
|
NM_000602
NM_001165413
|
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1
|
|
chrX_-_130250880
|
0.640
|
|
IGSF1
|
immunoglobulin superfamily, member 1
|
|
chr15_-_88148177
|
0.640
|
|
ANPEP
|
alanyl (membrane) aminopeptidase
|
|
chr15_-_56829069
|
0.637
|
|
ADAM10
|
ADAM metallopeptidase domain 10
|
|
chr6_+_37245860
|
0.633
|
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr17_+_24394281
|
0.630
|
|
PIPOX
|
pipecolic acid oxidase
|
|
chr2_+_8739563
|
0.628
|
NM_002166
|
ID2
|
inhibitor of DNA binding 2, dominant negative helix-loop-helix protein
|
|
chr6_+_31976754
|
0.627
|
NM_001178063
|
C2
|
complement component 2
|
|
chr7_+_73506333
|
0.627
|
NM_005685
|
GTF2IRD1
|
GTF2I repeat domain containing 1
|
|
chr4_-_39205316
|
0.626
|
|
UGDH
|
UDP-glucose 6-dehydrogenase
|