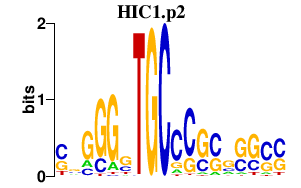
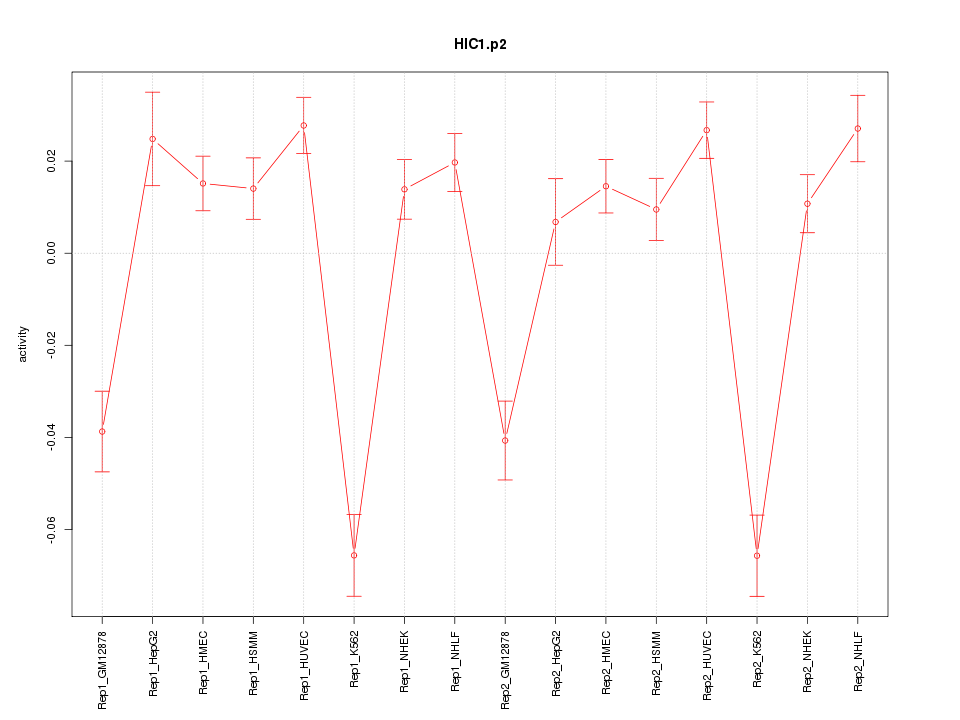
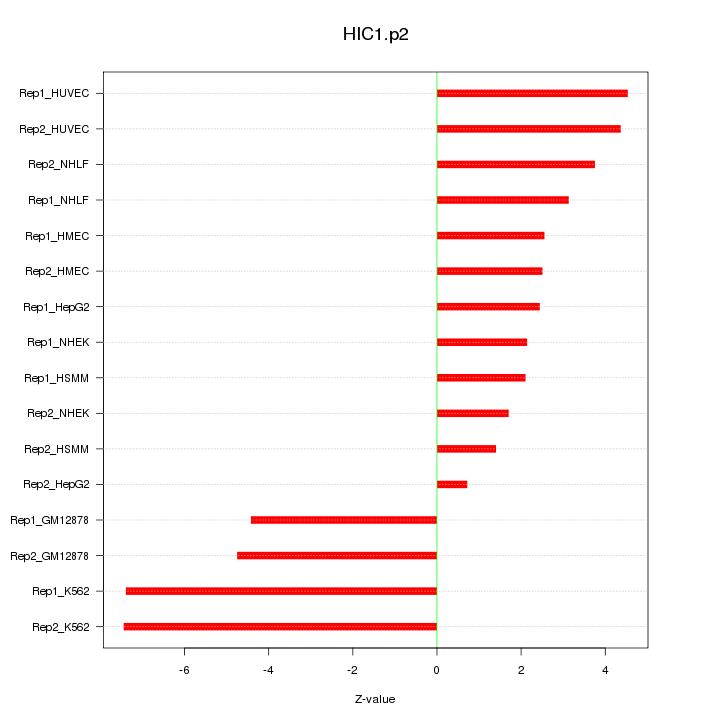
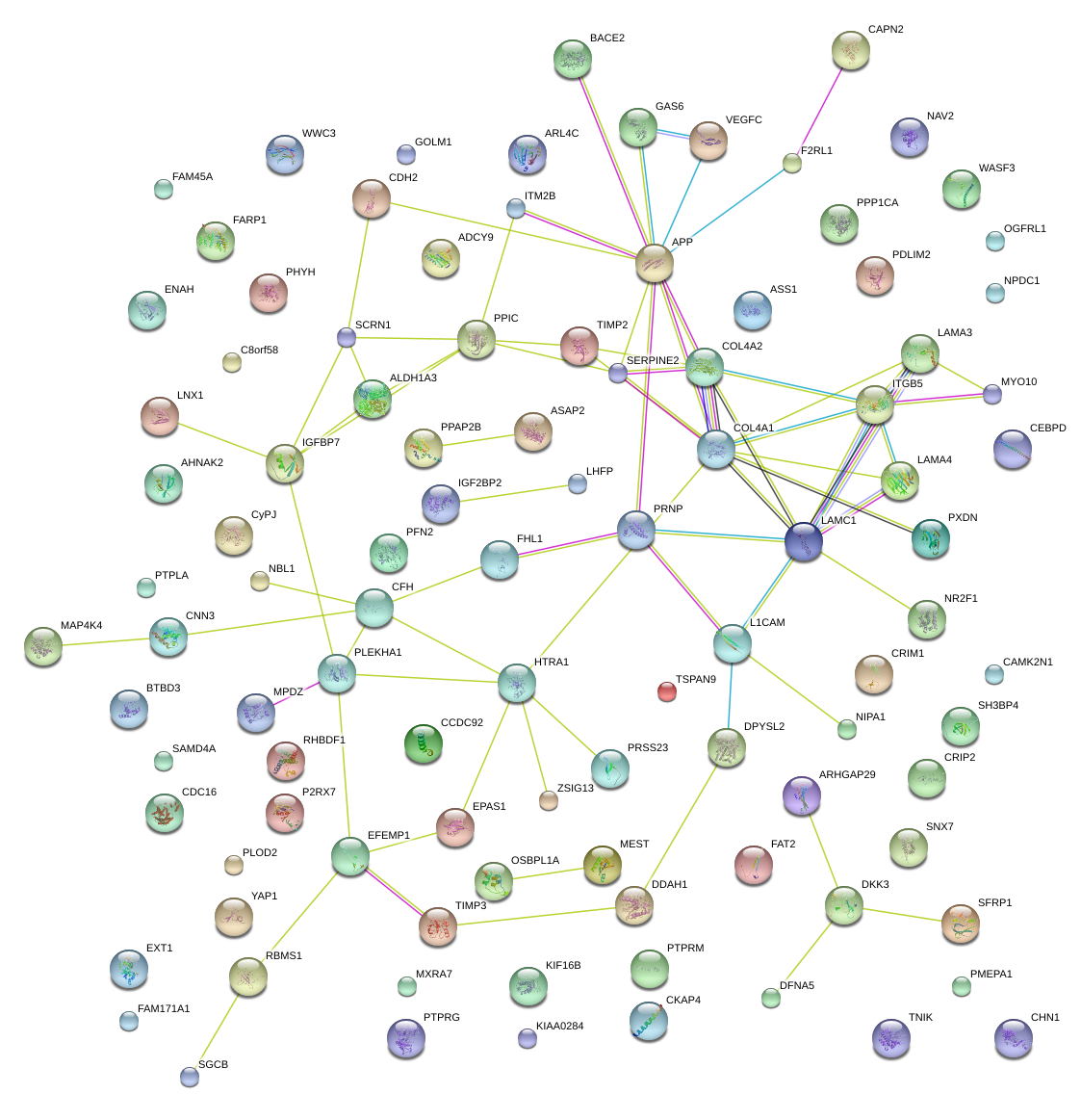
|
chr2_-_224612177
|
10.688
|
NM_001136528
NM_006216
|
SERPINE2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2
|
|
chr11_+_101485946
|
9.527
|
NM_001130145
NM_001195044
NM_006106
|
YAP1
|
Yes-associated protein 1
|
|
chr2_-_160972606
|
9.217
|
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr2_-_224611568
|
8.984
|
NM_001136530
|
SERPINE2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2
|
|
chr13_-_109757442
|
8.960
|
NM_001845
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr1_-_85703198
|
7.927
|
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr13_+_109757593
|
7.246
|
NM_001846
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr1_-_20685314
|
7.213
|
NM_018584
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr11_-_11987458
|
7.014
|
NM_013253
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr11_-_11987414
|
6.961
|
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr13_+_97593711
|
6.722
|
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr2_+_235525355
|
6.417
|
NM_014521
|
SH3BP4
|
SH3-domain binding protein 4
|
|
chr8_+_26491317
|
6.336
|
NM_001386
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr10_+_124124196
|
6.270
|
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr10_+_124211353
|
6.165
|
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr11_-_11987198
|
5.920
|
NM_015881
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr11_-_11987115
|
5.757
|
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr4_-_177950658
|
5.678
|
NM_005429
|
VEGFC
|
vascular endothelial growth factor C
|
|
chr2_+_9264327
|
5.670
|
NM_001135191
NM_003887
|
ASAP2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2
|
|
chr13_-_39075304
|
5.644
|
|
LHFP
|
lipoma HMGIC fusion partner
|
|
chr1_-_223907283
|
5.614
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr2_+_9264360
|
5.604
|
|
ASAP2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2
|
|
chr10_+_124210980
|
5.591
|
NM_002775
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr13_+_97593469
|
5.430
|
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr15_+_99237527
|
5.272
|
NM_000693
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3
|
|
chr3_-_187025501
|
5.267
|
NM_001007225
NM_006548
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr1_-_95165037
|
5.213
|
|
CNN3
|
calponin 3, acidic
|
|
chr12_-_123023052
|
5.183
|
NM_025140
|
CCDC92
|
coiled-coil domain containing 92
|
|
chr16_-_4106186
|
5.167
|
NM_001116
|
ADCY9
|
adenylate cyclase 9
|
|
chrX_+_9943793
|
5.098
|
NM_015691
|
WWC3
|
WWC family member 3
|
|
chr14_+_105012106
|
5.089
|
NM_001312
|
CRIP2
|
cysteine-rich protein 2
|
|
chr7_+_129919157
|
4.992
|
NM_002402
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr1_-_95165089
|
4.846
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr17_-_74433048
|
4.816
|
NM_003255
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr13_+_26029879
|
4.807
|
|
WASF3
|
WAS protein family, member 3
|
|
chr16_-_4106022
|
4.766
|
|
ADCY9
|
adenylate cyclase 9
|
|
chr2_-_235070431
|
4.730
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chr2_-_1727247
|
4.711
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr14_+_104402626
|
4.641
|
NM_001112726
NM_015005
|
KIAA0284
|
KIAA0284
|
|
chr11_+_86189138
|
4.637
|
NM_007173
|
PRSS23
|
protease, serine, 23
|
|
chr8_-_41286088
|
4.586
|
NM_003012
|
SFRP1
|
secreted frizzled-related protein 1
|
|
chr2_-_1727292
|
4.535
|
NM_012293
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr1_-_94475504
|
4.535
|
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr3_-_151171190
|
4.489
|
NM_002628
NM_053024
|
PFN2
|
profilin 2
|
|
chr13_+_113546943
|
4.460
|
|
GAS6
|
growth arrest-specific 6
|
|
chrX_+_135057202
|
4.458
|
NM_001159702
NM_001159703
NM_001449
|
FHL1
|
four and a half LIM domains 1
|
|
chr2_+_101680594
|
4.434
|
NM_004834
NM_145686
NM_145687
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr17_-_74432800
|
4.413
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr12_-_105165805
|
4.352
|
NM_006825
|
CKAP4
|
cytoskeleton-associated protein 4
|
|
chr1_+_19843261
|
4.320
|
NM_005380
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr1_-_94475840
|
4.307
|
NM_004815
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr8_+_22492586
|
4.274
|
|
PDLIM2
|
PDZ and LIM domain 2 (mystique)
|
|
chr18_+_7557313
|
4.273
|
NM_001105244
NM_002845
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chr9_-_13269562
|
4.271
|
|
MPDZ
|
multiple PDZ domain protein
|
|
chr9_-_87904306
|
4.237
|
NM_016548
|
GOLM1
|
golgi membrane protein 1
|
|
chr17_-_74432671
|
4.232
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr2_+_46377952
|
4.197
|
NM_001430
|
EPAS1
|
endothelial PAS domain protein 1
|
|
chr8_-_48813236
|
4.185
|
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta
|
|
chr2_-_161058120
|
4.162
|
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr1_+_181258952
|
4.132
|
NM_002293
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2)
|
|
chr13_+_113546841
|
4.088
|
NM_000820
|
GAS6
|
growth arrest-specific 6
|
|
chr2_-_175578156
|
4.069
|
NM_001025201
NM_001822
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr13_+_113546912
|
4.024
|
|
GAS6
|
growth arrest-specific 6
|
|
chrX_-_152804731
|
4.021
|
|
L1CAM
|
L1 cell adhesion molecule
|
|
chr21_+_41461962
|
4.013
|
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chr10_-_13381751
|
3.993
|
NM_001037537
|
PHYH
|
phytanoyl-CoA 2-hydroxylase
|
|
chr1_-_56817717
|
3.988
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr20_+_4614796
|
3.984
|
NM_000311
NM_001080121
NM_001080122
NM_183079
NM_001080123
|
PRNP
|
prion protein
|
|
chr8_-_119192985
|
3.950
|
NM_000127
|
EXT1
|
exostosin 1
|
|
chr1_-_56817823
|
3.948
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr13_-_39075355
|
3.930
|
NM_005780
|
LHFP
|
lipoma HMGIC fusion partner
|
|
chr10_-_17699371
|
3.929
|
NM_014241
|
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A
|
|
chr9_-_139060285
|
3.927
|
|
NPDC1
|
neural proliferation, differentiation and control, 1
|
|
chr10_-_17699309
|
3.884
|
|
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A
|
|
chr9_-_139060377
|
3.853
|
|
NPDC1
|
neural proliferation, differentiation and control, 1
|
|
chr1_-_56817568
|
3.847
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr13_-_39075200
|
3.840
|
|
LHFP
|
lipoma HMGIC fusion partner
|
|
chr7_-_24763556
|
3.812
|
|
DFNA5
|
deafness, autosomal dominant 5
|
|
chr13_+_113546897
|
3.794
|
|
GAS6
|
growth arrest-specific 6
|
|
chr13_+_26029799
|
3.789
|
NM_006646
|
WASF3
|
WAS protein family, member 3
|
|
chr1_-_85703321
|
3.757
|
NM_012137
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr6_+_72055235
|
3.738
|
|
OGFRL1
|
opioid growth factor receptor-like 1
|
|
chr10_+_124124083
|
3.718
|
NM_001001974
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr7_-_29995809
|
3.702
|
NM_001145515
NM_014766
NM_001145513
|
SCRN1
|
secernin 1
|
|
chr4_-_52599181
|
3.684
|
NM_000232
|
SGCB
|
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein)
|
|
chr3_-_126088819
|
3.663
|
NM_002213
|
ITGB5
|
integrin, beta 5
|
|
chr7_-_24763577
|
3.631
|
NM_001127453
|
DFNA5
|
deafness, autosomal dominant 5
|
|
chr22_+_31527678
|
3.588
|
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr5_+_92946348
|
3.575
|
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr5_-_16989371
|
3.565
|
NM_012334
|
MYO10
|
myosin X
|
|
chr11_+_19691397
|
3.490
|
NM_145117
NM_182964
|
NAV2
|
neuron navigator 2
|
|
chr21_+_45649719
|
3.416
|
|
|
|
|
chr13_+_47705493
|
3.394
|
|
ITM2B
|
integral membrane protein 2B
|
|
chr6_+_72055197
|
3.393
|
NM_024576
|
OGFRL1
|
opioid growth factor receptor-like 1
|
|
chr1_+_221966835
|
3.376
|
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr15_-_20637731
|
3.373
|
NM_144599
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1
|
|
chr7_+_129918408
|
3.327
|
NM_177525
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr3_-_147361465
|
3.318
|
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr15_+_99237555
|
3.310
|
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3
|
|
chr9_+_132309914
|
3.288
|
NM_000050
NM_054012
|
ASS1
|
argininosuccinate synthase 1
|
|
chr6_+_72055229
|
3.279
|
|
OGFRL1
|
opioid growth factor receptor-like 1
|
|
chr5_-_122400312
|
3.263
|
NM_000943
|
PPIC
|
peptidylprolyl isomerase C (cyclophilin C)
|
|
chr10_-_15453060
|
3.260
|
NM_001010924
|
FAM171A1
|
family with sequence similarity 171, member A1
|
|
chr5_+_76150588
|
3.241
|
NM_005242
|
F2RL1
|
coagulation factor II (thrombin) receptor-like 1
|
|
chr17_-_72218661
|
3.239
|
|
MXRA7
|
matrix-remodelling associated 7
|
|
chr21_-_26465005
|
3.231
|
NM_000484
NM_001136129
NM_001136130
NM_201413
NM_201414
|
APP
|
amyloid beta (A4) precursor protein
|
|
chr16_-_62573
|
3.210
|
NM_022450
|
RHBDF1
|
rhomboid 5 homolog 1 (Drosophila)
|
|
chr9_-_139060416
|
3.182
|
NM_015392
|
NPDC1
|
neural proliferation, differentiation and control, 1
|
|
chr4_-_57671112
|
3.177
|
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr3_+_61522219
|
3.159
|
NM_002841
|
PTPRG
|
protein tyrosine phosphatase, receptor type, G
|
|
chr1_+_98899734
|
3.137
|
NM_015976
NM_152238
|
SNX7
|
sorting nexin 7
|
|
chr18_-_20231709
|
3.077
|
|
OSBPL1A
|
oxysterol binding protein-like 1A
|
|
chr9_-_87904215
|
3.077
|
|
GOLM1
|
golgi membrane protein 1
|
|
chr9_-_87904280
|
3.069
|
|
GOLM1
|
golgi membrane protein 1
|
|
chr22_+_31527744
|
3.054
|
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr17_-_72218535
|
3.051
|
NM_001008528
NM_001008529
NM_198530
|
MXRA7
|
matrix-remodelling associated 7
|
|
chr7_-_24763841
|
3.038
|
|
DFNA5
|
deafness, autosomal dominant 5
|
|
chr14_+_54104383
|
3.028
|
NM_001161576
NM_015589
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr18_+_19523526
|
3.023
|
NM_001127717
NM_198129
|
LAMA3
|
laminin, alpha 3
|
|
chr21_+_41461924
|
3.022
|
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chr18_-_24011349
|
3.012
|
NM_001792
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr2_-_1727324
|
2.994
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr12_+_3056781
|
2.975
|
NM_001168320
NM_006675
|
TSPAN9
|
tetraspanin 9
|
|
chr2_-_56004366
|
2.962
|
NM_001039348
NM_001039349
|
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1
|
|
chr2_+_36436873
|
2.946
|
NM_016441
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr20_+_11819364
|
2.896
|
NM_181443
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr14_-_104515738
|
2.886
|
NM_138420
|
AHNAK2
|
AHNAK nucleoprotein 2
|
|
chr8_-_48813256
|
2.873
|
NM_005195
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta
|
|
chr10_+_120853548
|
2.863
|
|
FAM45A
|
family with sequence similarity 45, member A
|
|
chr8_+_26491383
|
2.863
|
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr20_-_55718351
|
2.842
|
NM_020182
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr12_-_123023312
|
2.830
|
|
CCDC92
|
coiled-coil domain containing 92
|
|
chr21_+_45649429
|
2.828
|
NM_130445
|
COL18A1
|
collagen, type XVIII, alpha 1
|
|
chr2_-_112907577
|
2.791
|
NM_001164463
|
RGPD8
RGPD5
|
RANBP2-like and GRIP domain containing 8
RANBP2-like and GRIP domain containing 5
|
|
chr3_-_187025438
|
2.782
|
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr7_+_79602023
|
2.754
|
NM_002069
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1
|
|
chr8_-_27528105
|
2.739
|
|
CLU
|
clusterin
|
|
chr21_+_41461597
|
2.725
|
NM_012105
NM_138991
NM_138992
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chr15_+_94674849
|
2.716
|
NM_021005
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr18_-_20105814
|
2.700
|
|
OSBPL1A
|
oxysterol binding protein-like 1A
|
|
chr3_-_126257425
|
2.672
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr10_+_120853600
|
2.669
|
NM_207009
|
FAM45A
|
family with sequence similarity 45, member A
|
|
chr1_+_85819004
|
2.661
|
NM_001554
|
CYR61
|
cysteine-rich, angiogenic inducer, 61
|
|
chr1_-_56817802
|
2.639
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr3_+_50167424
|
2.636
|
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F
|
|
chr15_-_20929658
|
2.634
|
|
HERC2P2
|
hect domain and RLD 2 pseudogene 2
|
|
chr9_+_123003549
|
2.628
|
|
GSN
|
gelsolin
|
|
chr2_+_173000552
|
2.626
|
NM_000210
NM_001079818
|
ITGA6
|
integrin, alpha 6
|
|
chr3_-_147451509
|
2.625
|
|
PLSCR4
|
phospholipid scramblase 4
|
|
chr5_-_122400219
|
2.617
|
|
PPIC
|
peptidylprolyl isomerase C (cyclophilin C)
|
|
chr12_+_6179736
|
2.605
|
NM_001769
|
CD9
|
CD9 molecule
|
|
chr8_-_17702665
|
2.599
|
NM_001001924
NM_001001925
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr19_+_16048266
|
2.597
|
|
TPM4
|
tropomyosin 4
|
|
chr1_+_167342149
|
2.587
|
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr3_-_147361912
|
2.566
|
NM_000935
NM_182943
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr12_-_108802561
|
2.540
|
NM_016433
|
GLTP
|
glycolipid transfer protein
|
|
chr11_+_129445017
|
2.538
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr4_-_22126437
|
2.532
|
NM_145290
|
GPR125
|
G protein-coupled receptor 125
|
|
chr11_+_129445057
|
2.497
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr3_-_151171576
|
2.496
|
|
PFN2
|
profilin 2
|
|
chr13_+_97593411
|
2.487
|
NM_001001715
NM_005766
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr3_-_147361626
|
2.471
|
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr5_+_76047536
|
2.467
|
NM_001992
|
F2R
|
coagulation factor II (thrombin) receptor
|
|
chr3_+_57969127
|
2.450
|
NM_001164317
NM_001164318
NM_001164319
NM_001457
|
FLNB
|
filamin B, beta
|
|
chr3_-_147451648
|
2.449
|
NM_001128306
NM_001177304
NM_020353
|
PLSCR4
|
phospholipid scramblase 4
|
|
chr5_-_57791544
|
2.449
|
NM_006622
|
PLK2
|
polo-like kinase 2
|
|
chr1_+_221966849
|
2.418
|
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr2_-_165406173
|
2.416
|
NM_014900
|
COBLL1
|
COBL-like 1
|
|
chr17_+_75689985
|
2.392
|
|
GAA
|
glucosidase, alpha; acid
|
|
chr15_-_81744469
|
2.389
|
NM_001717
|
BNC1
|
basonuclin 1
|
|
chr17_-_60088773
|
2.384
|
NM_022739
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2
|
|
chr17_-_60088459
|
2.376
|
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2
|
|
chr8_-_27528215
|
2.371
|
NM_001831
|
CLU
|
clusterin
|
|
chr18_-_20231783
|
2.363
|
NM_080597
|
OSBPL1A
|
oxysterol binding protein-like 1A
|
|
chr21_+_17807167
|
2.361
|
NM_001338
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr2_+_200879737
|
2.357
|
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like
|
|
chr19_-_4351470
|
2.348
|
NM_003025
|
SH3GL1
|
SH3-domain GRB2-like 1
|
|
chr7_-_150960086
|
2.344
|
NM_024429
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit
|
|
chr5_+_52812165
|
2.343
|
|
FST
|
follistatin
|
|
chr19_+_16048306
|
2.342
|
|
TPM4
|
tropomyosin 4
|
|
chr9_+_123070276
|
2.321
|
|
GSN
|
gelsolin
|
|
chr9_+_123070191
|
2.319
|
NM_001127662
NM_001127663
NM_001127664
NM_001127665
NM_001127666
NM_001127667
NM_198252
|
GSN
|
gelsolin
|
|
chr11_+_13255913
|
2.314
|
|
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like
|
|
chr3_-_49370755
|
2.312
|
NM_000581
NM_201397
|
GPX1
|
glutathione peroxidase 1
|
|
chr16_-_10581944
|
2.298
|
NM_001424
|
EMP2
|
epithelial membrane protein 2
|
|
chr14_-_51605461
|
2.290
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr17_-_829655
|
2.288
|
NM_022463
|
NXN
|
nucleoredoxin
|
|
chr5_-_146869811
|
2.274
|
NM_001197294
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr11_+_13255836
|
2.267
|
NM_001030272
NM_001030273
NM_001178
|
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like
|
|
chr4_+_184257337
|
2.266
|
NM_024949
|
WWC2
|
WW and C2 domain containing 2
|
|
chr12_-_107775462
|
2.263
|
NM_001161330
NM_018984
|
SSH1
|
slingshot homolog 1 (Drosophila)
|
|
chr5_+_76150637
|
2.262
|
|
F2RL1
|
coagulation factor II (thrombin) receptor-like 1
|
|
chr1_-_43691932
|
2.260
|
|
HYI
|
hydroxypyruvate isomerase (putative)
|
|
chr6_-_52549774
|
2.256
|
NM_012288
|
TRAM2
|
translocation associated membrane protein 2
|
|
chr14_+_85069217
|
2.247
|
|
|
|
|
chr18_-_490583
|
2.246
|
NM_130386
|
COLEC12
|
collectin sub-family member 12
|
|
chr11_-_2907169
|
2.239
|
NM_003311
|
PHLDA2
|
pleckstrin homology-like domain, family A, member 2
|
|
chr12_-_15833617
|
2.233
|
NM_004447
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr22_+_36928011
|
2.230
|
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr3_-_121652183
|
2.226
|
NM_007085
|
FSTL1
|
follistatin-like 1
|