|
chr11_+_12652427
|
3.686
|
NM_021961
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr20_+_19863716
|
3.080
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr6_+_139497933
|
3.056
|
NM_016217
|
HECA
|
headcase homolog (Drosophila)
|
|
chr19_+_16048266
|
2.971
|
|
TPM4
|
tropomyosin 4
|
|
chr19_+_16048315
|
2.852
|
|
TPM4
|
tropomyosin 4
|
|
chr19_+_16048306
|
2.660
|
|
TPM4
|
tropomyosin 4
|
|
chr2_-_216009015
|
2.562
|
NM_002026
NM_054034
NM_212474
NM_212476
NM_212478
NM_212482
|
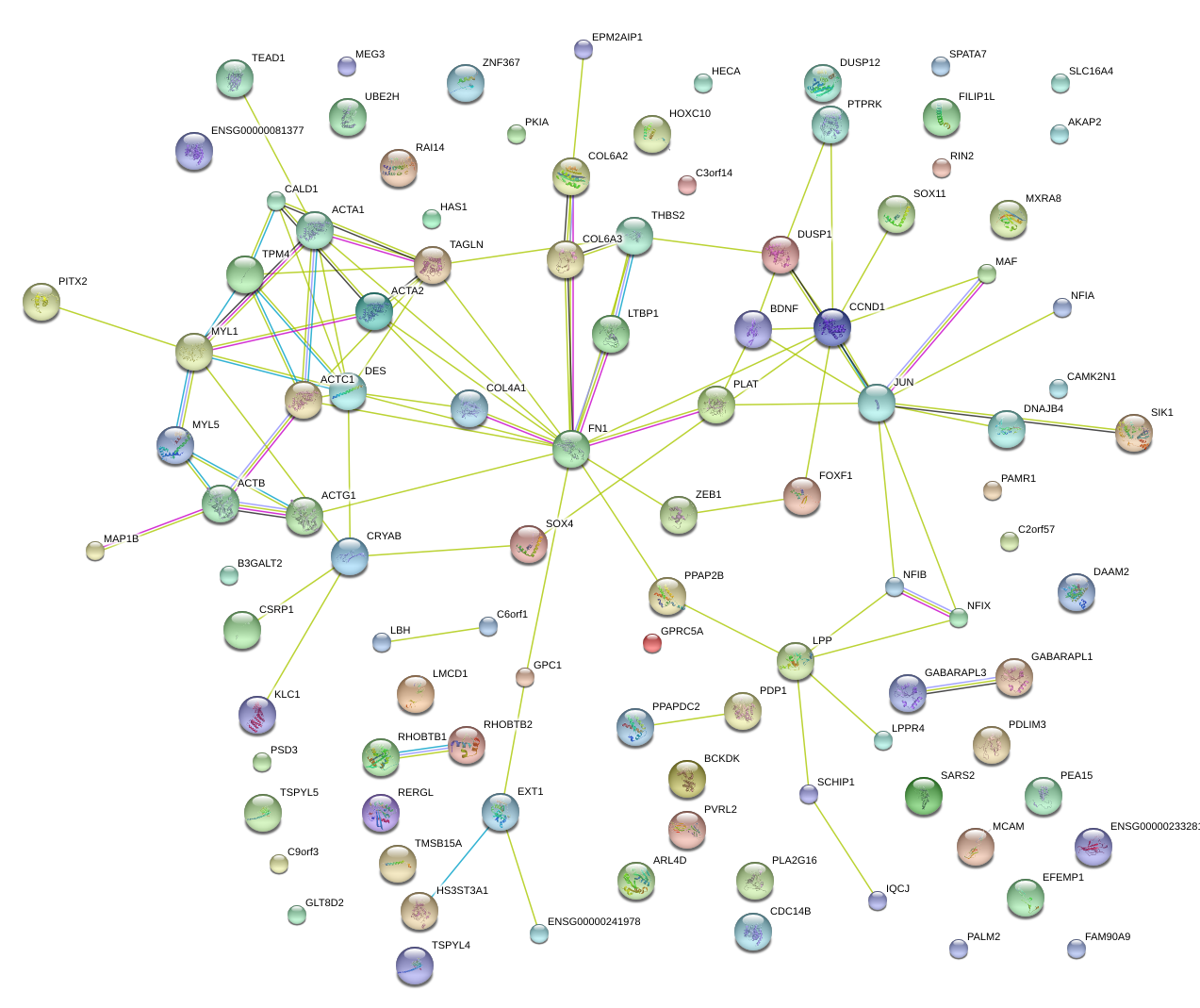
FN1
|
fibronectin 1
|
|
chr21_+_46369606
|
2.519
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr3_+_8518508
|
2.233
|
NM_014583
|
LMCD1
|
LIM and cysteine-rich domains 1
|
|
chr11_+_116575249
|
2.066
|
NM_001001522
NM_003186
|
TAGLN
|
transgelin
|
|
chr3_-_101077605
|
2.046
|
NM_014890
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr2_-_237987509
|
2.013
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr2_-_237987579
|
1.984
|
NM_004369
NM_057164
NM_057165
NM_057166
NM_057167
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr9_-_98421894
|
1.963
|
NM_003671
NM_033331
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr4_-_186693574
|
1.788
|
NM_001114107
NM_014476
|
PDLIM3
|
PDZ and LIM domain 3
|
|
chr3_+_8518539
|
1.748
|
|
LMCD1
|
LIM and cysteine-rich domains 1
|
|
chr4_-_111777651
|
1.695
|
|
PITX2
|
paired-like homeodomain 2
|
|
chr13_-_109600709
|
1.685
|
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr4_-_111777584
|
1.673
|
|
PITX2
|
paired-like homeodomain 2
|
|
chr5_-_172130766
|
1.638
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr2_-_237987554
|
1.549
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr2_-_56003859
|
1.524
|
NM_004105
|
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1
|
|
chr21_-_43671355
|
1.505
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr1_+_99502655
|
1.502
|
|
LPPR4
|
lipid phosphate phosphatase-related protein type 4
|
|
chr8_-_18710575
|
1.459
|
NM_206909
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr21_+_46370220
|
1.453
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr11_-_111287657
|
1.444
|
NM_001885
|
CRYAB
|
crystallin, alpha B
|
|
chr21_+_46369617
|
1.443
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr6_+_39868126
|
1.438
|
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr3_+_160473729
|
1.436
|
NM_001197107
NM_001197108
NM_014575
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr1_-_20685314
|
1.435
|
NM_018584
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr1_-_1283770
|
1.394
|
NM_032348
|
MXRA8
|
matrix-remodelling associated 8
|
|
chr1_-_59022275
|
1.379
|
NM_002228
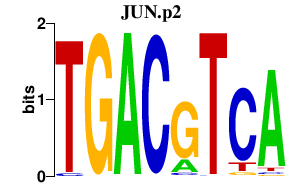
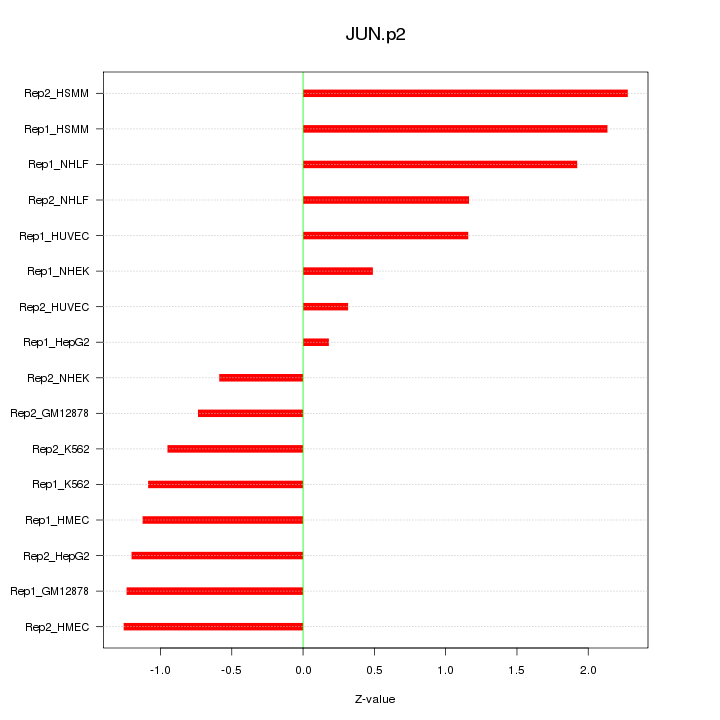
|
JUN
|
jun proto-oncogene
|
|
chr8_-_98359248
|
1.352
|
|
TSPYL5
|
TSPY-like 5
|
|
chr8_-_98359347
|
1.347
|
NM_033512
|
TSPYL5
|
TSPY-like 5
|
|
chr8_-_42184194
|
1.308
|
|
PLAT
|
plasminogen activator, tissue
|
|
chr8_-_42184350
|
1.306
|
NM_000930
NM_033011
|
PLAT
|
plasminogen activator, tissue
|
|
chr2_+_219991342
|
1.298
|
NM_001927
|
DES
|
desmin
|
|
chr11_-_35503720
|
1.285
|
NM_001001991
NM_015430
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr2_-_210876560
|
1.285
|
NM_079422
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast
|
|
chr8_+_79591074
|
1.256
|
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr11_+_69165119
|
1.255
|
|
CCND1
|
cyclin D1
|
|
chr11_+_69165018
|
1.242
|
NM_053056
|
CCND1
|
cyclin D1
|
|
chr21_+_46370831
|
1.237
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr8_+_94998258
|
1.217
|
NM_018444
NM_001161780
NM_001161779
NM_001161781
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1
|
|
chr1_-_56750235
|
1.167
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr19_+_16048047
|
1.155
|
NM_003290
|
TPM4
|
tropomyosin 4
|
|
chr3_+_189354167
|
1.137
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr4_-_111763655
|
1.135
|
NM_000325
|
PITX2
|
paired-like homeodomain 2
|
|
chr7_-_129379947
|
1.124
|
NM_003344
NM_182697
|
UBE2H
|
ubiquitin-conjugating enzyme E2H (UBC8 homolog, yeast)
|
|
chr14_+_103165244
|
1.119
|
NM_001130107
NM_005552
NM_182923
|
KLC1
|
kinesin light chain 1
|
|
chr1_-_191422346
|
1.114
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr2_+_232165818
|
1.112
|
NM_152614
|
C2orf57
|
chromosome 2 open reading frame 57
|
|
chr19_+_50041400
|
1.110
|
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr3_+_62279707
|
1.109
|
|
C3orf14
|
chromosome 3 open reading frame 14
|
|
chr5_+_71438796
|
1.084
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr4_+_658250
|
1.076
|
|
MYL5
|
myosin, light chain 5, regulatory
|
|
chr4_+_658312
|
1.060
|
|
MYL5
|
myosin, light chain 5, regulatory
|
|
chr2_+_30307900
|
1.059
|
NM_030915
|
LBH
|
limb bud and heart development homolog (mouse)
|
|
chr2_+_5750229
|
1.054
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr5_+_34723420
|
1.050
|
NM_001145523
|
RAI14
|
retinoic acid induced 14
|
|
chr10_+_31648103
|
1.045
|
NM_001174093
NM_001174095
NM_001174096
NM_030751
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr16_-_78192111
|
1.043
|
NM_001031804
NM_005360
|
MAF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian)
|
|
chrX_-_101658297
|
1.035
|
NM_021992
|
TMSB15A
TMSB15B
|
thymosin beta 15a
thymosin beta 15B
|
|
chr2_+_33025872
|
1.034
|
NM_206943
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr1_+_99502435
|
1.015
|
NM_001166252
NM_014839
|
LPPR4
|
lipid phosphate phosphatase-related protein type 4
|
|
chr16_+_85101605
|
1.013
|
NM_001451
|
FOXF1
|
forkhead box F1
|
|
chr6_-_169395847
|
1.008
|
|
THBS2
|
thrombospondin 2
|
|
chr17_-_13445942
|
0.997
|
NM_006042
|
HS3ST3A1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1
|
|
chr14_+_87921740
|
0.989
|
NM_001040428
NM_018418
|
SPATA7
|
spermatogenesis associated 7
|
|
chr2_-_216008689
|
0.982
|
|
FN1
|
fibronectin 1
|
|
chr12_+_10256682
|
0.977
|
NM_031412
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1
|
|
chr16_+_31027255
|
0.971
|
|
BCKDK
|
branched chain ketoacid dehydrogenase kinase
|
|
chr19_-_56919032
|
0.971
|
NM_001523
|
HAS1
|
hyaluronan synthase 1
|
|
chr9_+_111927601
|
0.968
|
NM_001136562
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr7_+_134114957
|
0.966
|
|
CALD1
|
caldesmon 1
|
|
chr19_+_50041387
|
0.959
|
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr12_+_52665196
|
0.958
|
NM_017409
|
HOXC10
|
homeobox C10
|
|
chr11_-_118693021
|
0.954
|
NM_006500
|
MCAM
|
melanoma cell adhesion molecule
|
|
chr1_-_110735158
|
0.950
|
NM_004696
|
SLC16A4
|
solute carrier family 16, member 4 (monocarboxylic acid transporter 5)
|
|
chr6_-_128883125
|
0.941
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr10_-_62373828
|
0.922
|
NM_014836
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chr1_+_78243181
|
0.915
|
NM_007034
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4
|
|
chr14_+_100362236
|
0.901
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr14_+_87921818
|
0.899
|
|
SPATA7
|
spermatogenesis associated 7
|
|
chr15_-_32875040
|
0.898
|
NM_005159
|
ACTC1
|
actin, alpha, cardiac muscle 1
|
|
chr17_+_38831874
|
0.885
|
NM_001661
|
ARL4D
|
ADP-ribosylation factor-like 4D
|
|
chr11_-_63138483
|
0.883
|
NM_007069
|
PLA2G16
|
phospholipase A2, group XVI
|
|
chr16_+_31027231
|
0.882
|
|
BCKDK
|
branched chain ketoacid dehydrogenase kinase
|
|
chr2_+_241023703
|
0.879
|
NM_002081
|
GPC1
|
glypican 1
|
|
chr9_+_96528814
|
0.878
|
|
C9orf3
|
chromosome 9 open reading frame 3
|
|
chr9_+_96528764
|
0.877
|
NM_001193331
NM_032823
|
C9orf3
|
chromosome 9 open reading frame 3
|
|
chr11_-_63138416
|
0.877
|
|
PLA2G16
|
phospholipase A2, group XVI
|
|
chr1_+_158441770
|
0.875
|
|
PEA15
|
phosphoprotein enriched in astrocytes 15
|
|
chr9_-_14076901
|
0.871
|
|
NFIB
|
nuclear factor I/B
|
|
chr11_-_27679617
|
0.867
|
NM_170733
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr12_+_12935619
|
0.864
|
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr14_+_100362204
|
0.854
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr1_+_158441738
|
0.853
|
NM_003768
|
PEA15
|
phosphoprotein enriched in astrocytes 15
|
|
chr1_-_199742864
|
0.852
|
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr8_-_119192985
|
0.852
|
NM_000127
|
EXT1
|
exostosin 1
|
|
chr3_+_11242666
|
0.836
|
NM_001098211
|
HRH1
|
histamine receptor H1
|
|
chr9_+_34980266
|
0.835
|
NM_001135005
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5
|
|
chr16_-_10581944
|
0.829
|
NM_001424
|
EMP2
|
epithelial membrane protein 2
|
|
chr1_-_199742871
|
0.826
|
NM_001193570
NM_004078
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr3_+_9950474
|
0.821
|
|
CRELD1
|
cysteine-rich with EGF-like domains 1
|
|
chr18_+_8707368
|
0.805
|
NM_015210
|
KIAA0802
|
KIAA0802
|
|
chr1_-_199742589
|
0.801
|
NM_001193571
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr3_+_156280129
|
0.800
|
NM_000902
|
MME
|
membrane metallo-endopeptidase
|
|
chr1_-_199742841
|
0.798
|
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr3_-_109423858
|
0.797
|
NM_018010
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas)
|
|
chrX_-_10605726
|
0.789
|
NM_001098624
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr22_+_36927884
|
0.788
|
NM_001161572
NM_001161574
NM_012323
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr11_-_63138516
|
0.783
|
NM_001128203
|
PLA2G16
|
phospholipase A2, group XVI
|
|
chr11_+_69165159
|
0.775
|
|
CCND1
|
cyclin D1
|
|
chr6_-_46401363
|
0.760
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chr17_+_64020120
|
0.760
|
NM_002734
NM_212472
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1)
|
|
chr17_-_61253122
|
0.755
|
NM_001037325
|
CCDC46
|
coiled-coil domain containing 46
|
|
chr1_+_24944493
|
0.754
|
|
CLIC4
|
chloride intracellular channel 4
|
|
chr10_+_102782156
|
0.745
|
|
SFXN3
|
sideroflexin 3
|
|
chr3_-_126257425
|
0.743
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr5_-_131590833
|
0.741
|
NM_001017973
NM_004199
|
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II
|
|
chr5_+_71538456
|
0.740
|
|
MAP1B
|
microtubule-associated protein 1B
|
|
chrX_-_13866451
|
0.738
|
|
GPM6B
|
glycoprotein M6B
|
|
chr17_+_64020159
|
0.737
|
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1)
|
|
chr9_-_128924819
|
0.737
|
NM_012098
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr9_-_128924706
|
0.732
|
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr19_+_50041424
|
0.728
|
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr10_+_102782151
|
0.728
|
|
SFXN3
|
sideroflexin 3
|
|
chr2_+_28469199
|
0.722
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr3_-_188946168
|
0.719
|
NM_001706
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr12_-_100979923
|
0.716
|
NM_016053
|
CCDC53
|
coiled-coil domain containing 53
|
|
chr22_+_36928011
|
0.714
|
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr2_-_86704446
|
0.714
|
|
RNF103
|
ring finger protein 103
|
|
chrX_+_153325684
|
0.708
|
|
FAM50A
|
family with sequence similarity 50, member A
|
|
chr4_-_171161486
|
0.693
|
NM_001009554
|
MFAP3L
|
microfibrillar-associated protein 3-like
|
|
chr12_+_12935759
|
0.683
|
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr1_+_24944346
|
0.679
|
NM_013943
|
CLIC4
|
chloride intracellular channel 4
|
|
chr13_-_51878629
|
0.675
|
NM_018676
NM_199263
|
THSD1
|
thrombospondin, type I, domain containing 1
|
|
chrX_+_48317772
|
0.665
|
NM_006743
|
RBM3
|
RNA binding motif (RNP1, RRM) protein 3
|
|
chr1_+_7753913
|
0.664
|
NM_004781
|
VAMP3
|
vesicle-associated membrane protein 3 (cellubrevin)
|
|
chr7_+_65307816
|
0.663
|
|
TPST1
|
tyrosylprotein sulfotransferase 1
|
|
chr7_+_65307691
|
0.662
|
NM_003596
|
TPST1
|
tyrosylprotein sulfotransferase 1
|
|
chr9_+_96561750
|
0.660
|
NM_001193329
|
C9orf3
|
chromosome 9 open reading frame 3
|
|
chr6_-_112682573
|
0.655
|
|
LAMA4
|
laminin, alpha 4
|
|
chr8_-_18585721
|
0.654
|
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr7_+_142790047
|
0.651
|
|
ZYX
|
zyxin
|
|
chr9_+_111582317
|
0.651
|
NM_053016
NM_007203
NM_147150
|
PALM2
PALM2-AKAP2
|
paralemmin 2
PALM2-AKAP2 readthrough
|
|
chr17_+_22645232
|
0.646
|
NM_015626
NM_134265
|
WSB1
|
WD repeat and SOCS box containing 1
|
|
chr20_+_19818209
|
0.642
|
NM_018993
|
RIN2
|
Ras and Rab interactor 2
|
|
chr10_+_123913094
|
0.640
|
NM_006997
NM_206860
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr3_+_9950513
|
0.638
|
NM_015513
NM_001031717
NM_001077415
|
CRELD1
|
cysteine-rich with EGF-like domains 1
|
|
chr14_+_95792299
|
0.636
|
NM_000710
|
BDKRB1
|
bradykinin receptor B1
|
|
chr6_+_148705421
|
0.627
|
NM_015278
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr7_-_37922902
|
0.620
|
|
SFRP4
|
secreted frizzled-related protein 4
|
|
chr12_+_120635021
|
0.620
|
NM_001080825
|
TMEM120B
|
transmembrane protein 120B
|
|
chr6_+_148705646
|
0.617
|
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr4_-_156517394
|
0.615
|
|
MAP9
|
microtubule-associated protein 9
|
|
chrX_+_153325643
|
0.610
|
NM_004699
|
FAM50A
|
family with sequence similarity 50, member A
|
|
chr1_+_24944554
|
0.610
|
|
CLIC4
|
chloride intracellular channel 4
|
|
chr6_+_148705817
|
0.605
|
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr21_-_43903755
|
0.605
|
NM_007031
|
HSF2BP
|
heat shock transcription factor 2 binding protein
|
|
chr12_-_100979893
|
0.600
|
|
CCDC53
|
coiled-coil domain containing 53
|
|
chr21_+_46369911
|
0.600
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr7_+_142788579
|
0.599
|
|
ZYX
|
zyxin
|
|
chr6_-_116681863
|
0.598
|
NM_021648
|
TSPYL4
|
TSPY-like 4
|
|
chr19_-_63758270
|
0.595
|
|
CHMP2A
|
chromatin modifying protein 2A
|
|
chr3_+_101694244
|
0.586
|
|
TMEM45A
|
transmembrane protein 45A
|
|
chr12_-_100979869
|
0.585
|
|
CCDC53
|
coiled-coil domain containing 53
|
|
chr17_+_64020155
|
0.585
|
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1)
|
|
chr7_-_45095017
|
0.585
|
NM_001146334
|
NACAD
|
NAC alpha domain containing
|
|
chr7_+_142788521
|
0.583
|
|
ZYX
|
zyxin
|
|
chr19_-_63758184
|
0.582
|
|
CHMP2A
|
chromatin modifying protein 2A
|
|
chrX_+_12903145
|
0.581
|
NM_021109
NM_183049
|
TMSB4X
TMSL3
|
thymosin beta 4, X-linked
thymosin-like 3
|
|
chr1_-_95165089
|
0.580
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr11_+_129445002
|
0.579
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr7_-_37923040
|
0.575
|
NM_003014
|
SFRP4
|
secreted frizzled-related protein 4
|
|
chr10_+_123912930
|
0.574
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr7_+_142788730
|
0.567
|
|
ZYX
|
zyxin
|
|
chr22_-_37062114
|
0.565
|
|
CSNK1E
|
casein kinase 1, epsilon
|
|
chr19_-_63758289
|
0.563
|
NM_014453
|
CHMP2A
|
chromatin modifying protein 2A
|
|
chr12_-_121946635
|
0.556
|
NM_024667
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae)
|
|
chr13_+_47705493
|
0.552
|
|
ITM2B
|
integral membrane protein 2B
|
|
chr7_+_142788480
|
0.551
|
NM_001010972
NM_003461
|
ZYX
|
zyxin
|
|
chr7_+_142788566
|
0.551
|
|
ZYX
|
zyxin
|
|
chr7_+_142788517
|
0.547
|
|
ZYX
|
zyxin
|
|
chr12_+_55201854
|
0.546
|
NM_002898
|
RBMS2
|
RNA binding motif, single stranded interacting protein 2
|
|
chr7_+_142788569
|
0.545
|
|
ZYX
|
zyxin
|
|
chr19_-_40938251
|
0.544
|
|
HSPB6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chr4_-_156517423
|
0.541
|
|
MAP9
|
microtubule-associated protein 9
|
|
chrX_-_13866565
|
0.540
|
NM_001001994
|
GPM6B
|
glycoprotein M6B
|
|
chr20_+_42644633
|
0.539
|
|
PKIG
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma
|
|
chr11_+_129445017
|
0.537
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr4_+_146622154
|
0.536
|
NM_005900
|
SMAD1
|
SMAD family member 1
|
|
chr19_-_63758165
|
0.534
|
NM_198426
|
CHMP2A
|
chromatin modifying protein 2A
|
|
chrX_-_92815222
|
0.532
|
NM_004538
|
NAP1L3
|
nucleosome assembly protein 1-like 3
|
|
chr21_+_34112456
|
0.531
|
|
ITSN1
|
intersectin 1 (SH3 domain protein)
|
|
chr2_-_224175339
|
0.530
|
|
SCG2
|
secretogranin II
|
|
chr21_-_38210531
|
0.526
|
NM_002240
|
KCNJ6
|
potassium inwardly-rectifying channel, subfamily J, member 6
|
|
chr1_+_156229686
|
0.525
|
NM_018240
|
KIRREL
|
kin of IRRE like (Drosophila)
|