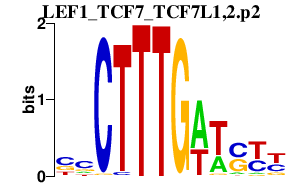
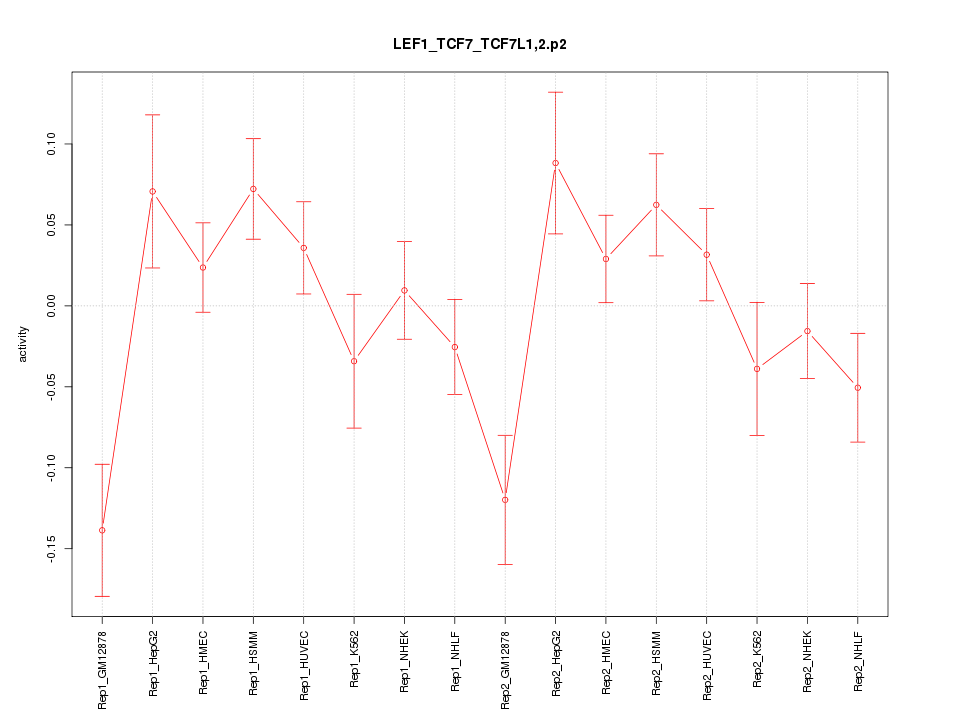
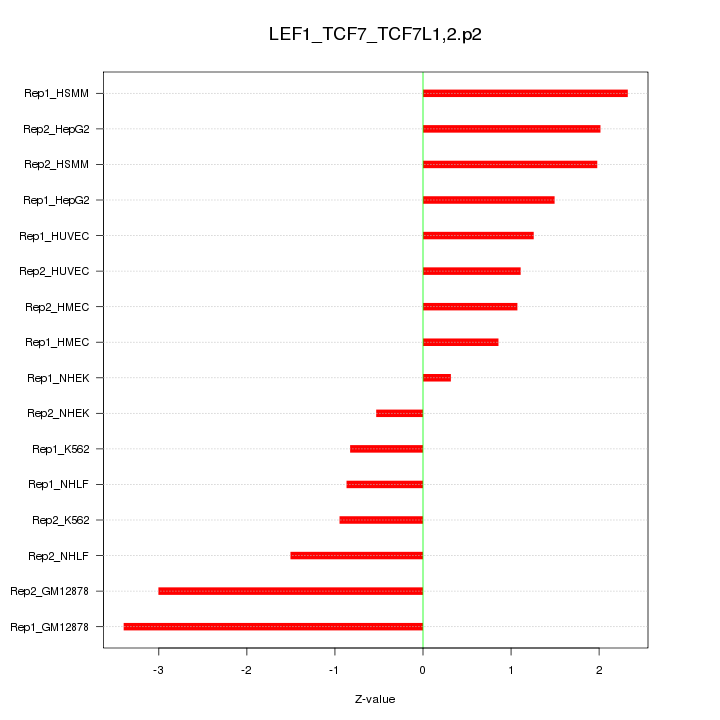
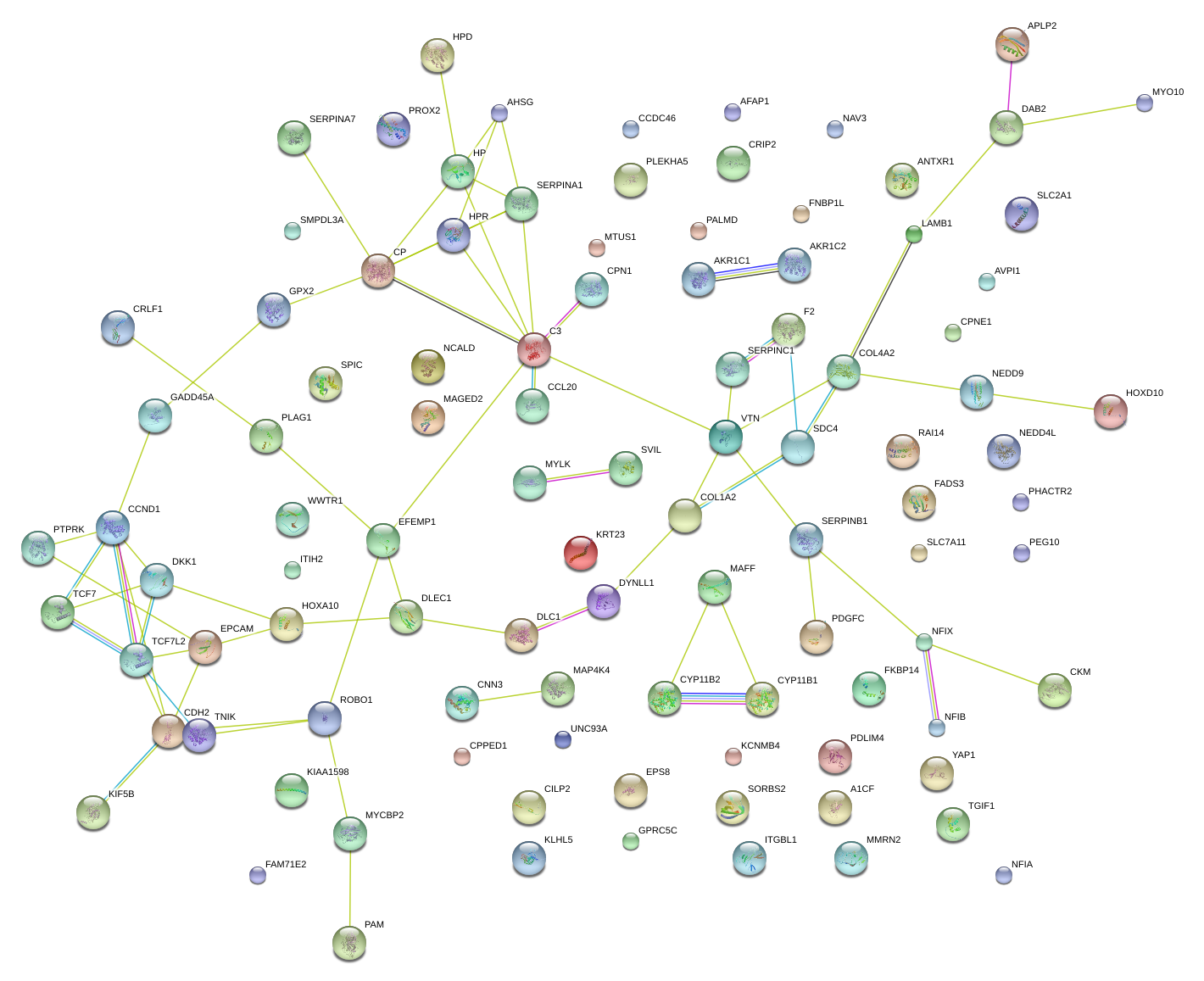
|
chr10_+_53744043
|
5.774
|
NM_012242
|
DKK1
|
dickkopf homolog 1 (Xenopus laevis)
|
|
chr6_-_128883125
|
5.357
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr10_+_7785347
|
3.761
|
|
ITIH2
|
inter-alpha (globulin) inhibitor H2
|
|
chr4_-_187114407
|
3.748
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr10_+_7785360
|
3.534
|
|
ITIH2
|
inter-alpha (globulin) inhibitor H2
|
|
chr10_+_7785331
|
3.490
|
|
ITIH2
|
inter-alpha (globulin) inhibitor H2
|
|
chr18_-_24011349
|
3.469
|
NM_001792
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr11_+_69165119
|
2.985
|
|
CCND1
|
cyclin D1
|
|
chr11_+_69165018
|
2.943
|
NM_053056
|
CCND1
|
cyclin D1
|
|
chr10_+_7785235
|
2.787
|
NM_002216
|
ITIH2
|
inter-alpha (globulin) inhibitor H2
|
|
chr1_+_93686154
|
2.682
|
NM_001024948
NM_001164473
NM_017737
|
FNBP1L
|
formin binding protein 1-like
|
|
chr3_-_150858274
|
2.597
|
NM_001168280
NM_015472
|
WWTR1
|
WW domain containing transcription regulator 1
|
|
chr7_-_107430566
|
2.482
|
NM_002291
|
LAMB1
|
laminin, beta 1
|
|
chr5_-_16989371
|
2.479
|
NM_012334
|
MYO10
|
myosin X
|
|
chr10_+_7785290
|
2.444
|
|
ITIH2
|
inter-alpha (globulin) inhibitor H2
|
|
chr19_-_18578577
|
2.400
|
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr19_-_18578467
|
2.351
|
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr5_+_34720368
|
2.335
|
NM_001145521
NM_001145525
|
RAI14
|
retinoic acid induced 14
|
|
chr4_-_158111995
|
2.324
|
NM_016205
|
PDGFC
|
platelet derived growth factor C
|
|
chrX_-_105169370
|
2.297
|
NM_000354
|
SERPINA7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7
|
|
chr5_-_39430180
|
2.246
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr19_-_18578631
|
2.219
|
NM_004750
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr5_-_16791346
|
2.168
|
|
MYO10
|
myosin X
|
|
chr12_-_120781002
|
2.165
|
NM_002150
|
HPD
|
4-hydroxyphenylpyruvate dioxygenase
|
|
chr2_+_101680594
|
2.079
|
NM_004834
NM_145686
NM_145687
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr18_+_3439972
|
2.070
|
NM_173209
NM_173208
NM_003244
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr10_+_114699953
|
1.979
|
NM_001146274
NM_001146283
NM_001146284
NM_001146285
NM_001146286
NM_001198525
NM_001198526
NM_001198527
NM_001198528
NM_001198529
NM_001198530
NM_001198531
NM_030756
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr10_+_4995453
|
1.950
|
NM_001353
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr4_+_38740246
|
1.929
|
NM_015990
NM_199039
|
KLHL5
|
kelch-like 5 (Drosophila)
|
|
chr4_-_158111504
|
1.916
|
|
PDGFC
|
platelet derived growth factor C
|
|
chr2_+_228386800
|
1.852
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr13_+_109935250
|
1.807
|
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr18_-_24010944
|
1.769
|
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr10_-_118755077
|
1.730
|
NM_001127211
NM_018330
|
KIAA1598
|
KIAA1598
|
|
chr11_+_46697345
|
1.723
|
|
F2
|
coagulation factor II (thrombin)
|
|
chr11_+_69165159
|
1.680
|
|
CCND1
|
cyclin D1
|
|
chr11_+_46697312
|
1.663
|
NM_000506
|
F2
|
coagulation factor II (thrombin)
|
|
chr18_+_54039779
|
1.625
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr3_-_150422474
|
1.578
|
NM_000096
|
CP
|
ceruloplasmin (ferroxidase)
|
|
chr14_-_64479198
|
1.577
|
NM_002083
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal)
|
|
chr5_+_131621287
|
1.541
|
|
PDLIM4
|
PDZ and LIM domain 4
|
|
chr13_+_109932934
|
1.529
|
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr11_+_101488380
|
1.500
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr18_+_54039818
|
1.483
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr6_+_143971009
|
1.469
|
NM_001100166
NM_014721
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr2_+_47449951
|
1.447
|
|
EPCAM
|
epithelial cell adhesion molecule
|
|
chr9_-_14388981
|
1.421
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr7_-_27186400
|
1.389
|
NM_153715
|
HOXA10
|
homeobox A10
|
|
chr14_+_105012106
|
1.383
|
NM_001312
|
CRIP2
|
cysteine-rich protein 2
|
|
chr22_-_34566367
|
1.377
|
NM_001031695
NM_001082576
NM_001082577
NM_014309
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr12_-_15833383
|
1.372
|
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr8_-_17624009
|
1.330
|
NM_001001931
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr4_-_139382672
|
1.308
|
NM_014331
|
SLC7A11
|
solute carrier family 7, (cationic amino acid transporter, y+ system) member 11
|
|
chr19_-_6671585
|
1.300
|
NM_000064
|
C3
|
complement component 3
|
|
chr16_-_12805194
|
1.293
|
NM_001099455
NM_018340
|
CPPED1
|
calcineurin-like phosphoesterase domain containing 1
|
|
chr3_-_124822113
|
1.293
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr1_-_95165089
|
1.269
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr22_+_36928972
|
1.268
|
NM_001161573
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr7_+_94130673
|
1.256
|
|
PEG10
|
paternally expressed 10
|
|
chr12_+_19173669
|
1.229
|
NM_001143821
NM_001190860
NM_019012
|
PLEKHA5
|
pleckstrin homology domain containing, family A member 5
|
|
chr6_-_11340886
|
1.227
|
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr6_-_2785682
|
1.225
|
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1
|
|
chrX_+_54852217
|
1.212
|
NM_201222
|
MAGED2
|
melanoma antigen family D, 2
|
|
chr5_-_39460687
|
1.209
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr7_+_94130752
|
1.207
|
|
PEG10
|
paternally expressed 10
|
|
chr17_-_36347197
|
1.202
|
NM_015515
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr3_-_124821868
|
1.201
|
|
MYLK
|
myosin light chain kinase
|
|
chr13_+_109936080
|
1.196
|
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr11_+_69165065
|
1.191
|
|
CCND1
|
cyclin D1
|
|
chr17_-_61253122
|
1.168
|
NM_001037325
|
CCDC46
|
coiled-coil domain containing 46
|
|
chr20_-_43410437
|
1.165
|
NM_002999
|
SDC4
|
syndecan 4
|
|
chr4_-_7991992
|
1.129
|
NM_001134647
NM_198595
|
AFAP1
|
actin filament associated protein 1
|
|
chr18_+_54039730
|
1.122
|
NM_001144966
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr6_+_123151669
|
1.120
|
NM_006714
|
SMPDL3A
|
sphingomyelin phosphodiesterase, acid-like 3A
|
|
chr7_-_30032649
|
1.112
|
NM_017946
|
FKBP14
|
FK506 binding protein 14, 22 kDa
|
|
chr2_+_69093641
|
1.098
|
NM_018153
NM_032208
NM_053034
|
ANTXR1
|
anthrax toxin receptor 1
|
|
chr10_-_32385343
|
1.081
|
NM_004521
|
KIF5B
|
kinesin family member 5B
|
|
chr2_-_56003859
|
1.071
|
NM_004105
|
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1
|
|
chr14_-_93924663
|
1.067
|
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr1_-_43196911
|
1.067
|
|
SLC2A1
|
solute carrier family 2 (facilitated glucose transporter), member 1
|
|
chr10_-_52315390
|
1.065
|
|
A1CF
|
APOBEC1 complementation factor
|
|
chr19_-_50506883
|
1.060
|
|
CKM
|
creatine kinase, muscle
|
|
chr1_+_99884018
|
1.052
|
NM_017734
|
PALMD
|
palmdelphin
|
|
chr10_-_99436889
|
1.034
|
NM_021732
|
AVPI1
|
arginine vasopressin-induced 1
|
|
chr5_+_131621276
|
1.034
|
|
PDLIM4
|
PDZ and LIM domain 4
|
|
chr17_+_69939806
|
1.028
|
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C
|
|
chr3_-_79899748
|
1.022
|
NM_002941
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr17_-_23721468
|
1.022
|
|
VTN
|
vitronectin
|
|
chr5_+_102229425
|
1.022
|
NM_000919
NM_001177306
NM_138766
NM_138821
NM_138822
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr6_-_11340869
|
1.013
|
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr10_-_32385299
|
1.008
|
|
KIF5B
|
kinesin family member 5B
|
|
chr11_-_61415429
|
1.008
|
NM_021727
|
FADS3
|
fatty acid desaturase 3
|
|
chr8_-_17624304
|
1.007
|
NM_001166393
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr5_+_131621230
|
1.006
|
NM_001131027
NM_003687
|
PDLIM4
|
PDZ and LIM domain 4
|
|
chr6_+_167624792
|
1.004
|
NM_001143947
NM_018974
|
UNC93A
|
unc-93 homolog A (C. elegans)
|
|
chr1_+_67923660
|
1.003
|
|
GADD45A
|
growth arrest and DNA-damage-inducible, alpha
|
|
chr2_+_47449790
|
0.994
|
NM_002354
|
EPCAM
|
epithelial cell adhesion molecule
|
|
chr6_-_11340887
|
0.989
|
NM_006403
NM_182966
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr12_+_69046241
|
0.982
|
NM_014505
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4
|
|
chr12_+_76749199
|
0.982
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr13_+_100902942
|
0.975
|
NM_004791
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains)
|
|
chr10_-_88707299
|
0.948
|
NM_024756
|
MMRN2
|
multimerin 2
|
|
chr3_+_187813405
|
0.944
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr10_-_30064600
|
0.944
|
NM_003174
|
SVIL
|
supervillin
|
|
chr5_-_39461054
|
0.925
|
NM_001343
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr7_+_93862137
|
0.919
|
|
COL1A2
PLAG1
|
collagen, type I, alpha 2
pleiomorphic adenoma gene 1
|
|
chr16_+_70646008
|
0.868
|
NM_001126102
NM_005143
|
HP
|
haptoglobin
|
|
chr1_-_172153087
|
0.853
|
NM_000488
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1
|
|
chr10_-_101831579
|
0.849
|
NM_001308
|
CPN1
|
carboxypeptidase N, polypeptide 1
|
|
chr11_+_129484540
|
0.844
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr7_-_27180398
|
0.833
|
NM_018951
|
HOXA10
|
homeobox A10
|
|
chr11_-_88864061
|
0.831
|
NM_001143836
NM_016931
|
NOX4
|
NADPH oxidase 4
|
|
chr17_-_53849929
|
0.828
|
NM_017763
|
RNF43
|
ring finger protein 43
|
|
chr10_-_32385129
|
0.815
|
|
KIF5B
|
kinesin family member 5B
|
|
chr3_-_115960807
|
0.815
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr5_-_58370693
|
0.811
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr18_+_3437607
|
0.809
|
NM_173207
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr14_-_104515738
|
0.801
|
NM_138420
|
AHNAK2
|
AHNAK nucleoprotein 2
|
|
chr7_-_139523187
|
0.797
|
NM_030647
|
JHDM1D
|
jumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae)
|
|
chr2_+_219354948
|
0.795
|
|
CYP27A1
|
cytochrome P450, family 27, subfamily A, polypeptide 1
|
|
chr10_-_52315436
|
0.794
|
NM_001198818
NM_001198819
NM_001198820
NM_014576
NM_138932
NM_138933
|
A1CF
|
APOBEC1 complementation factor
|
|
chr15_+_22772545
|
0.777
|
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N
|
|
chr4_-_40911212
|
0.758
|
NM_001166050
NM_004307
NM_173075
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr14_-_73555566
|
0.757
|
NM_001249
|
ENTPD5
|
ectonucleoside triphosphate diphosphohydrolase 5
|
|
chr9_-_98185815
|
0.746
|
|
SLC35D2
|
solute carrier family 35, member D2
|
|
chr5_+_36642424
|
0.746
|
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr20_+_11846536
|
0.743
|
NM_014962
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr7_+_29200636
|
0.740
|
NM_004067
|
CHN2
|
chimerin (chimaerin) 2
|
|
chr7_+_73506333
|
0.740
|
NM_005685
|
GTF2IRD1
|
GTF2I repeat domain containing 1
|
|
chr7_+_93861808
|
0.734
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr15_-_88148538
|
0.730
|
|
ANPEP
|
alanyl (membrane) aminopeptidase
|
|
chr6_-_32253819
|
0.726
|
NM_032741
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha)
|
|
chr3_+_161040343
|
0.723
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr21_+_34112456
|
0.721
|
|
ITSN1
|
intersectin 1 (SH3 domain protein)
|
|
chr17_-_53849888
|
0.711
|
|
RNF43
|
ring finger protein 43
|
|
chr3_+_161040439
|
0.701
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr5_+_109053034
|
0.700
|
NM_002372
|
MAN2A1
|
mannosidase, alpha, class 2A, member 1
|
|
chr4_+_69996780
|
0.699
|
NM_001074
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7
|
|
chr7_+_73506172
|
0.691
|
|
GTF2IRD1
|
GTF2I repeat domain containing 1
|
|
chr12_-_15833617
|
0.689
|
NM_004447
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr6_-_47361921
|
0.686
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr11_+_86189138
|
0.682
|
NM_007173
|
PRSS23
|
protease, serine, 23
|
|
chr7_-_133794404
|
0.676
|
NM_001628
|
AKR1B1
|
aldo-keto reductase family 1, member B1 (aldose reductase)
|
|
chr14_-_20340830
|
0.673
|
NM_002933
NM_198234
NM_198235
|
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic)
|
|
chr6_-_134680888
|
0.672
|
NM_001143676
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr9_-_98185613
|
0.668
|
NM_007001
|
SLC35D2
|
solute carrier family 35, member D2
|
|
chr2_-_39517795
|
0.668
|
|
MAP4K3
|
mitogen-activated protein kinase kinase kinase kinase 3
|
|
chr1_-_43197123
|
0.665
|
|
SLC2A1
|
solute carrier family 2 (facilitated glucose transporter), member 1
|
|
chr8_-_11763037
|
0.648
|
NM_001908
NM_147780
NM_147781
NM_147782
NM_147783
|
CTSB
|
cathepsin B
|
|
chr5_+_66160359
|
0.646
|
NM_015183
|
MAST4
|
microtubule associated serine/threonine kinase family member 4
|
|
chr7_+_73506047
|
0.640
|
NM_016328
|
GTF2IRD1
|
GTF2I repeat domain containing 1
|
|
chr3_-_135575925
|
0.639
|
NM_016201
|
AMOTL2
|
angiomotin like 2
|
|
chr11_-_88962426
|
0.638
|
NM_001143837
|
NOX4
|
NADPH oxidase 4
|
|
chr10_-_116434364
|
0.636
|
NM_001003407
NM_001003408
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr17_-_60088637
|
0.628
|
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2
|
|
chr5_-_114543632
|
0.627
|
NM_001017397
NM_001017398
NM_018700
|
TRIM36
|
tripartite motif containing 36
|
|
chr5_-_39460787
|
0.627
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr11_-_65138217
|
0.625
|
NM_002419
|
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11
|
|
chr14_+_85066207
|
0.616
|
NM_013231
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chrX_+_135057202
|
0.614
|
NM_001159702
NM_001159703
NM_001449
|
FHL1
|
four and a half LIM domains 1
|
|
chr6_+_123152044
|
0.614
|
|
SMPDL3A
|
sphingomyelin phosphodiesterase, acid-like 3A
|
|
chr11_+_44073674
|
0.612
|
NM_001178083
NM_207122
|
EXT2
|
exostosin 2
|
|
chr11_-_72063059
|
0.612
|
NM_002599
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated
|
|
chr20_+_9146036
|
0.609
|
NM_182797
|
PLCB4
|
phospholipase C, beta 4
|
|
chr10_+_73393998
|
0.608
|
NM_004273
|
CHST3
|
carbohydrate (chondroitin 6) sulfotransferase 3
|
|
chrX_-_13744933
|
0.605
|
|
GPM6B
|
glycoprotein M6B
|
|
chr10_+_102722275
|
0.604
|
NM_017893
|
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G
|
|
chr15_-_86890796
|
0.601
|
NM_001144074
NM_017996
|
DET1
|
de-etiolated homolog 1 (Arabidopsis)
|
|
chr5_+_36642213
|
0.600
|
NM_001166696
NM_004172
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr10_+_4995685
|
0.584
|
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr8_+_32698892
|
0.583
|
NM_001159996
|
NRG1
|
neuregulin 1
|
|
chr10_-_5035967
|
0.581
|
NM_001135241
|
AKR1C1
AKR1C2
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III)
|
|
chrX_+_55951280
|
0.581
|
|
|
|
|
chr16_-_18720192
|
0.576
|
NM_015161
|
ARL6IP1
|
ADP-ribosylation factor-like 6 interacting protein 1
|
|
chr13_-_32658215
|
0.573
|
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr17_-_36346240
|
0.571
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr6_-_11887265
|
0.570
|
NM_001143948
NM_032744
|
C6orf105
|
chromosome 6 open reading frame 105
|
|
chr12_-_88573949
|
0.566
|
NM_001001323
NM_001682
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1
|
|
chr1_+_170768914
|
0.565
|
NM_016227
|
C1orf9
|
chromosome 1 open reading frame 9
|
|
chr12_-_120810899
|
0.565
|
NM_001171993
|
HPD
|
4-hydroxyphenylpyruvate dioxygenase
|
|
chr16_+_56259657
|
0.564
|
NM_170776
|
GPR97
|
G protein-coupled receptor 97
|
|
chr4_-_111777956
|
0.563
|
NM_153426
NM_153427
|
PITX2
|
paired-like homeodomain 2
|
|
chr4_+_146622154
|
0.562
|
NM_005900
|
SMAD1
|
SMAD family member 1
|
|
chr17_+_19377732
|
0.559
|
NM_018242
|
SLC47A1
|
solute carrier family 47, member 1
|
|
chr2_+_9264360
|
0.556
|
|
ASAP2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2
|
|
chrX_+_46822751
|
0.554
|
|
RGN
|
regucalcin (senescence marker protein-30)
|
|
chr14_+_85066265
|
0.550
|
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr1_+_98899734
|
0.547
|
NM_015976
NM_152238
|
SNX7
|
sorting nexin 7
|
|
chr14_+_94148466
|
0.543
|
NM_001085
|
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3
|
|
chr4_-_80079393
|
0.535
|
NM_001040202
|
PAQR3
|
progestin and adipoQ receptor family member III
|
|
chr9_+_494661
|
0.533
|
NM_015158
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chrX_+_102518764
|
0.531
|
NM_014380
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr20_-_45410033
|
0.529
|
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chrX_+_46822688
|
0.526
|
NM_004683
NM_152869
|
RGN
|
regucalcin (senescence marker protein-30)
|
|
chr2_-_86418143
|
0.526
|
NM_001164731
NM_001164732
NM_022912
|
REEP1
|
receptor accessory protein 1
|
|
chr19_+_18357966
|
0.524
|
NM_004864
|
GDF15
|
growth differentiation factor 15
|
|
chrX_+_70359780
|
0.522
|
NM_000166
|
GJB1
|
gap junction protein, beta 1, 32kDa
|
|
chr4_-_70661018
|
0.517
|
NM_014465
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1
|
|
chr12_-_6321417
|
0.515
|
NM_001065
|
TNFRSF1A
|
tumor necrosis factor receptor superfamily, member 1A
|
|
chr17_+_54997667
|
0.507
|
NM_001166301
NM_024612
|
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40
|