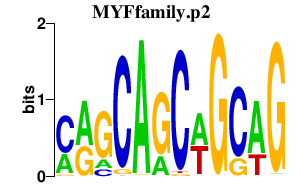
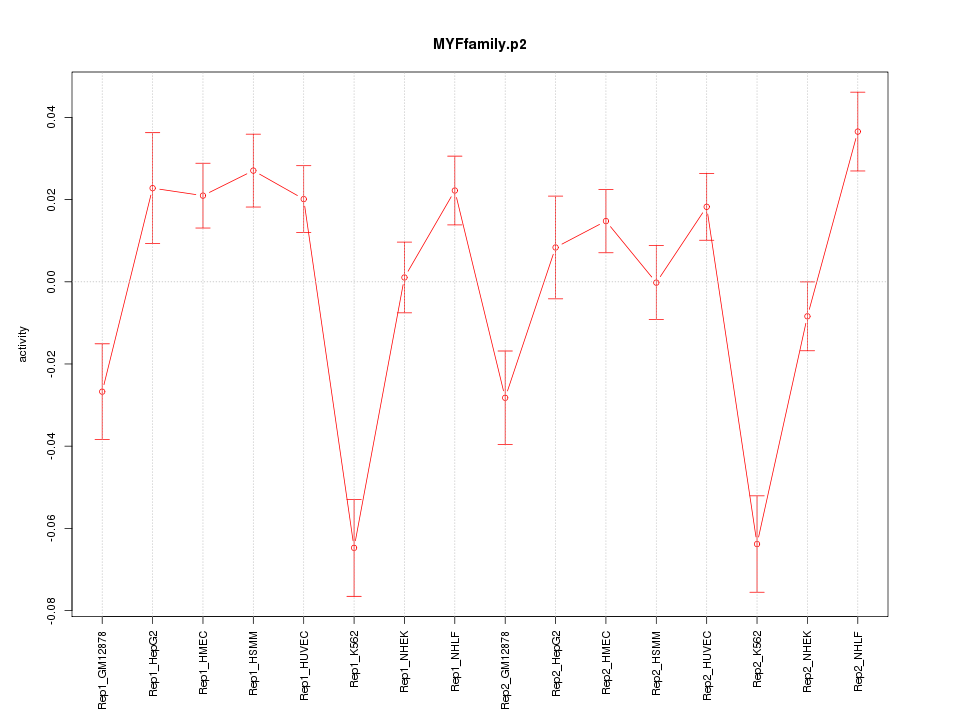
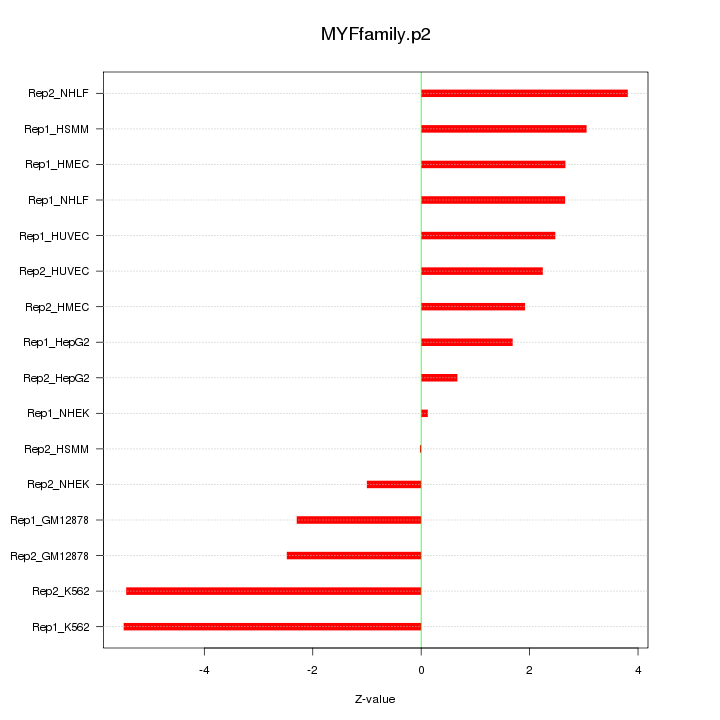
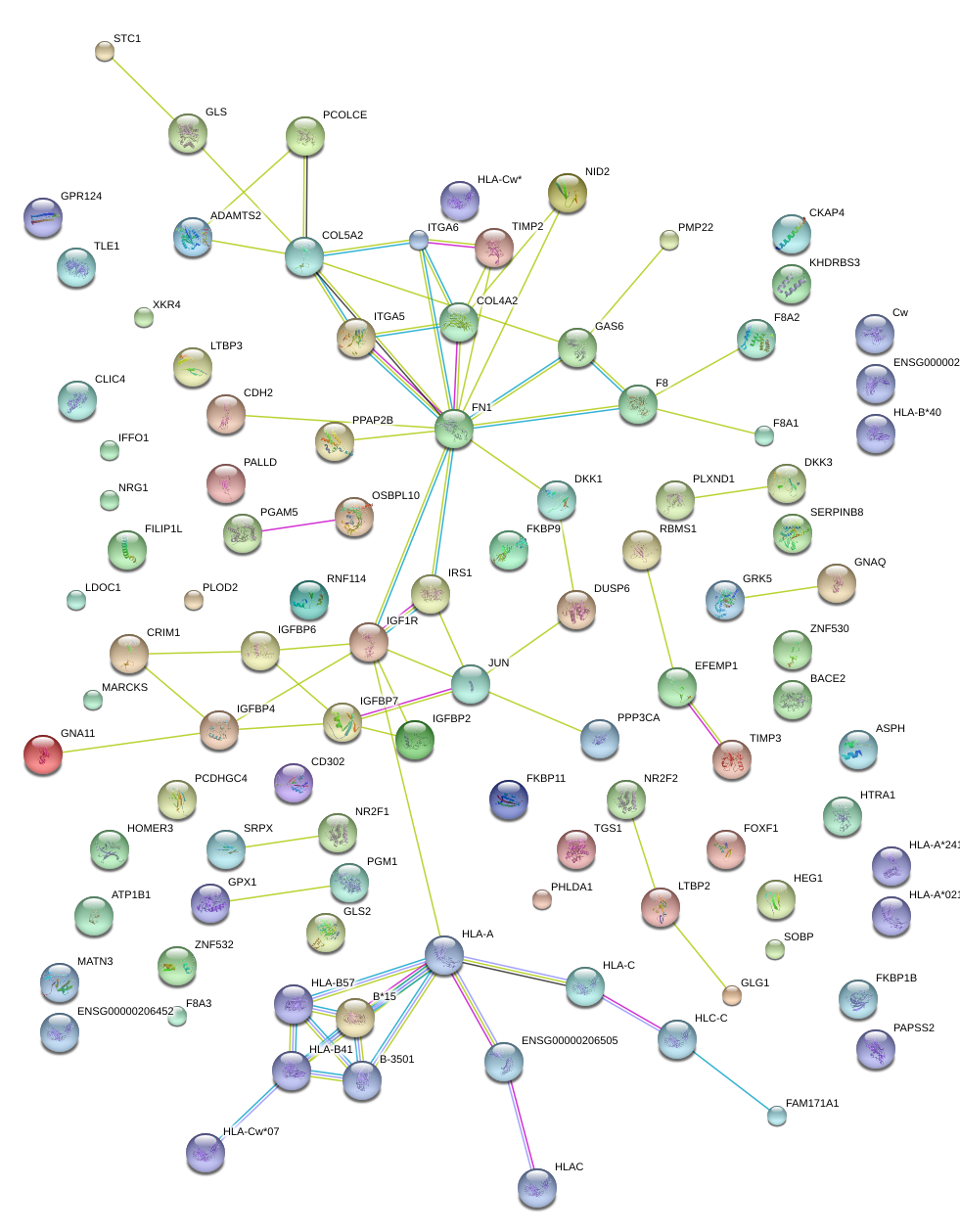
|
chr3_-_31997738
|
8.985
|
|
OSBPL10
|
oxysterol binding protein-like 10
|
|
chr12_-_74710741
|
8.415
|
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr4_-_57671273
|
8.136
|
NM_001553
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr4_-_57671307
|
7.983
|
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr12_-_74711474
|
7.557
|
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr4_-_57671112
|
6.854
|
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr1_-_56817802
|
6.491
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr2_-_227372574
|
6.171
|
|
IRS1
|
insulin receptor substrate 1
|
|
chr12_-_105165805
|
5.726
|
NM_006825
|
CKAP4
|
cytoskeleton-associated protein 4
|
|
chr12_-_74711642
|
5.708
|
NM_007350
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr10_+_124210980
|
4.982
|
NM_002775
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr1_-_56817717
|
4.958
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr2_-_161058120
|
4.854
|
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr16_-_73198416
|
4.820
|
|
GLG1
|
golgi glycoprotein 1
|
|
chr12_-_88270128
|
4.673
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr8_+_56177567
|
4.464
|
NM_052898
|
XKR4
|
XK, Kell blood group complex subunit-related family, member 4
|
|
chr13_+_113546912
|
4.429
|
|
GAS6
|
growth arrest-specific 6
|
|
chr6_+_107917965
|
4.366
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chrX_-_37964924
|
4.219
|
NM_001170750
NM_001170751
NM_001170752
NM_006307
|
SRPX
|
sushi-repeat containing protein, X-linked
|
|
chr13_+_113546943
|
4.076
|
|
GAS6
|
growth arrest-specific 6
|
|
chr1_-_56817568
|
4.050
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr10_+_89409332
|
4.029
|
NM_001015880
NM_004670
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2
|
|
chr13_+_113546841
|
3.834
|
NM_000820
|
GAS6
|
growth arrest-specific 6
|
|
chr2_+_36436873
|
3.753
|
NM_016441
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr2_+_217206450
|
3.720
|
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa
|
|
chr13_+_113546897
|
3.692
|
|
GAS6
|
growth arrest-specific 6
|
|
chr2_-_161058550
|
3.613
|
NM_002897
NM_016836
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr1_+_167342149
|
3.519
|
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr2_-_160362982
|
3.313
|
NM_001198763
NM_001198764
NM_014880
|
CD302
|
CD302 molecule
|
|
chr8_+_31616809
|
3.272
|
NM_013962
|
NRG1
|
neuregulin 1
|
|
chr14_-_51605487
|
3.265
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr3_-_147361465
|
3.134
|
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr3_-_147361626
|
3.130
|
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr2_+_217206327
|
3.100
|
NM_000597
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa
|
|
chr2_-_160362914
|
3.087
|
|
CD302
|
CD302 molecule
|
|
chr4_+_169989680
|
3.052
|
NM_001166110
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr3_-_49370682
|
3.019
|
|
GPX1
|
glutathione peroxidase 1
|
|
chr1_-_59020875
|
3.016
|
|
JUN
|
jun proto-oncogene
|
|
chr6_-_31347867
|
3.010
|
|
HLA-C
|
major histocompatibility complex, class I, C
|
|
chr2_-_56004366
|
2.976
|
NM_001039348
NM_001039349
|
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1
|
|
chr10_+_124211353
|
2.975
|
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr6_+_114285209
|
2.945
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr3_-_126257425
|
2.882
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr11_-_11986704
|
2.859
|
NM_001018057
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chrX_+_153767803
|
2.760
|
NM_001007523
NM_001007524
NM_012151
|
F8A2
F8A3
F8A1
|
coagulation factor VIII-associated (intronic transcript) 2
coagulation factor VIII-associated (intronic transcript) 3
coagulation factor VIII-associated (intronic transcript) 1
|
|
chr18_+_59788242
|
2.750
|
NM_001031848
NM_002640
NM_198833
|
SERPINB8
|
serpin peptidase inhibitor, clade B (ovalbumin), member 8
|
|
chr8_-_23768242
|
2.725
|
NM_003155
|
STC1
|
stanniocalcin 1
|
|
chrX_-_154341451
|
2.623
|
NM_001007523
NM_001007524
NM_012151
|
F8A2
F8A3
F8A1
|
coagulation factor VIII-associated (intronic transcript) 2
coagulation factor VIII-associated (intronic transcript) 3
coagulation factor VIII-associated (intronic transcript) 1
|
|
chr5_+_140844810
|
2.607
|
NM_018928
NM_032406
|
PCDHGC4
|
protocadherin gamma subfamily C, 4
|
|
chr13_+_109757593
|
2.579
|
NM_001846
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr2_-_227371698
|
2.567
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr16_-_73198498
|
2.537
|
|
GLG1
|
golgi glycoprotein 1
|
|
chr14_-_74148422
|
2.535
|
NM_000428
|
LTBP2
|
latent transforming growth factor beta binding protein 2
|
|
chr2_-_160972606
|
2.517
|
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr6_-_31347805
|
2.512
|
NM_002117
|
HLA-C
|
major histocompatibility complex, class I, C
|
|
chr16_-_73198515
|
2.488
|
NM_001145666
NM_001145667
NM_012201
|
GLG1
|
golgi glycoprotein 1
|
|
chr22_+_31527678
|
2.467
|
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr17_-_74433048
|
2.433
|
NM_003255
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr9_-_83494198
|
2.385
|
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr2_-_20075889
|
2.379
|
|
MATN3
|
matrilin 3
|
|
chr2_+_191454036
|
2.336
|
|
GLS
|
glutaminase
|
|
chr19_+_3045266
|
2.334
|
NM_002067
|
GNA11
|
guanine nucleotide binding protein (G protein), alpha 11 (Gq class)
|
|
chr5_+_92946348
|
2.256
|
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr2_+_173000552
|
2.241
|
NM_000210
NM_001079818
|
ITGA6
|
integrin, alpha 6
|
|
chr17_-_74432800
|
2.234
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr8_+_37773991
|
2.224
|
|
GPR124
|
G protein-coupled receptor 124
|
|
chr19_+_3045528
|
2.220
|
|
GNA11
|
guanine nucleotide binding protein (G protein), alpha 11 (Gq class)
|
|
chr10_-_15453060
|
2.211
|
NM_001010924
|
FAM171A1
|
family with sequence similarity 171, member A1
|
|
chr10_+_53744043
|
2.207
|
NM_012242
|
DKK1
|
dickkopf homolog 1 (Xenopus laevis)
|
|
chr1_+_63831479
|
2.206
|
NM_002633
|
PGM1
|
phosphoglucomutase 1
|
|
chr2_-_189752575
|
2.180
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr21_+_41461962
|
2.174
|
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chr1_+_24944346
|
2.169
|
NM_013943
|
CLIC4
|
chloride intracellular channel 4
|
|
chr12_-_47605478
|
2.107
|
NM_001143782
NM_016594
|
FKBP11
|
FK506 binding protein 11, 19 kDa
|
|
chr15_+_94674849
|
2.103
|
NM_021005
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr8_+_37773564
|
2.099
|
|
GPR124
|
G protein-coupled receptor 124
|
|
chr8_+_37773553
|
2.019
|
NM_032777
|
GPR124
|
G protein-coupled receptor 124
|
|
chr10_+_120957084
|
2.003
|
NM_005308
|
GRK5
|
G protein-coupled receptor kinase 5
|
|
chr15_+_97009194
|
1.979
|
|
IGF1R
|
insulin-like growth factor 1 receptor
|
|
chr2_+_191453791
|
1.967
|
NM_014905
|
GLS
|
glutaminase
|
|
chr4_-_102487356
|
1.964
|
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme
|
|
chr18_+_54681657
|
1.948
|
|
ZNF532
|
zinc finger protein 532
|
|
chr17_-_15104768
|
1.944
|
NM_153322
|
PMP22
|
peripheral myelin protein 22
|
|
chr12_-_53099269
|
1.938
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr3_-_130807865
|
1.935
|
NM_015103
|
PLXND1
|
plexin D1
|
|
chr2_+_173000590
|
1.917
|
|
ITGA6
|
integrin, alpha 6
|
|
chr1_+_24944493
|
1.905
|
|
CLIC4
|
chloride intracellular channel 4
|
|
chr2_-_216009015
|
1.896
|
NM_002026
NM_054034
NM_212474
NM_212476
NM_212478
NM_212482
|
FN1
|
fibronectin 1
|
|
chr21_+_41461597
|
1.879
|
NM_012105
NM_138991
NM_138992
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chr19_-_18912112
|
1.873
|
NM_001145722
NM_001145721
|
HOMER3
|
homer homolog 3 (Drosophila)
|
|
chrX_-_140098941
|
1.869
|
|
LDOC1
|
leucine zipper, down-regulated in cancer 1
|
|
chr18_-_24010944
|
1.865
|
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr17_+_35853177
|
1.862
|
NM_001552
|
IGFBP4
|
insulin-like growth factor binding protein 4
|
|
chrX_-_140098959
|
1.857
|
NM_012317
|
LDOC1
|
leucine zipper, down-regulated in cancer 1
|
|
chr12_-_6535481
|
1.837
|
NM_001039670
NM_001193457
NM_080730
|
IFFO1
|
intermediate filament family orphan 1
|
|
chr8_+_136539290
|
1.836
|
|
KHDRBS3
|
KH domain containing, RNA binding, signal transduction associated 3
|
|
chr5_-_178705036
|
1.817
|
NM_014244
NM_021599
|
ADAMTS2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2
|
|
chr8_+_37773915
|
1.813
|
|
GPR124
|
G protein-coupled receptor 124
|
|
chr8_-_62789665
|
1.798
|
|
ASPH
|
aspartate beta-hydroxylase
|
|
chr18_-_24011349
|
1.790
|
NM_001792
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr8_-_62789483
|
1.779
|
|
ASPH
|
aspartate beta-hydroxylase
|
|
chr7_+_32963486
|
1.774
|
NM_007270
|
FKBP9
|
FK506 binding protein 9, 63 kDa
|
|
chrX_-_140099014
|
1.758
|
|
LDOC1
|
leucine zipper, down-regulated in cancer 1
|
|
chrX_-_37916138
|
1.752
|
|
SRPX
|
sushi-repeat containing protein, X-linked
|
|
chr3_-_101316038
|
1.752
|
NM_001042459
NM_182909
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr16_+_85101605
|
1.746
|
NM_001451
|
FOXF1
|
forkhead box F1
|
|
chr12_+_51777684
|
1.744
|
NM_002178
|
IGFBP6
|
insulin-like growth factor binding protein 6
|
|
chr7_+_100037817
|
1.731
|
NM_002593
|
PCOLCE
|
procollagen C-endopeptidase enhancer
|
|
chr11_+_32069183
|
1.716
|
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain
|
|
chr12_-_7153068
|
1.715
|
NM_016546
|
C1RL
|
complement component 1, r subcomponent-like
|
|
chr2_+_191453813
|
1.709
|
|
GLS
|
glutaminase
|
|
chr6_-_132314004
|
1.706
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr4_-_102486975
|
1.687
|
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme
|
|
chr3_-_79151232
|
1.687
|
NM_001145845
NM_133631
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr22_-_35011703
|
1.687
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr22_-_35113797
|
1.670
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr20_-_23014828
|
1.665
|
NM_012072
|
CD93
|
CD93 molecule
|
|
chr7_+_93861808
|
1.644
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr13_-_109236897
|
1.641
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr12_-_53099203
|
1.630
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr21_+_45699830
|
1.623
|
NM_030582
NM_130444
|
COL18A1
|
collagen, type XVIII, alpha 1
|
|
chr19_-_54835162
|
1.622
|
NM_006270
|
RRAS
|
related RAS viral (r-ras) oncogene homolog
|
|
chr22_+_29819343
|
1.601
|
|
SMTN
|
smoothelin
|
|
chr20_+_3399615
|
1.585
|
NM_139321
NM_139322
|
ATRN
|
attractin
|
|
chr4_-_120769275
|
1.566
|
NM_001083
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific
|
|
chr11_-_6298284
|
1.562
|
NM_145040
|
PRKCDBP
|
protein kinase C, delta binding protein
|
|
chr1_+_24944554
|
1.555
|
|
CLIC4
|
chloride intracellular channel 4
|
|
chr3_-_147361912
|
1.540
|
NM_000935
NM_182943
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr12_-_53099277
|
1.539
|
NM_002205
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr1_-_20682541
|
1.537
|
|
|
|
|
chr9_-_14304036
|
1.534
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr4_-_102487649
|
1.515
|
NM_000944
NM_001130691
NM_001130692
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme
|
|
chr6_+_43151922
|
1.499
|
NM_002821
NM_152880
NM_152881
NM_152882
|
PTK7
|
PTK7 protein tyrosine kinase 7
|
|
chr2_-_220144427
|
1.494
|
|
OBSL1
|
obscurin-like 1
|
|
chr2_-_21120420
|
1.480
|
NM_000384
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr11_-_86343859
|
1.479
|
NM_012193
|
FZD4
|
frizzled homolog 4 (Drosophila)
|
|
chr21_-_27139143
|
1.467
|
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr12_+_51777715
|
1.457
|
|
IGFBP6
|
insulin-like growth factor binding protein 6
|
|
chr8_+_17478214
|
1.448
|
NM_006207
|
PDGFRL
|
platelet-derived growth factor receptor-like
|
|
chr5_-_178704934
|
1.440
|
|
ADAMTS2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2
|
|
chr3_-_151171576
|
1.417
|
|
PFN2
|
profilin 2
|
|
chr2_+_71148915
|
1.414
|
NM_017567
|
NAGK
|
N-acetylglucosamine kinase
|
|
chr8_-_62789721
|
1.402
|
NM_001164754
NM_001164755
NM_001164756
NM_004318
NM_032466
|
ASPH
|
aspartate beta-hydroxylase
|
|
chr9_+_123070276
|
1.398
|
|
GSN
|
gelsolin
|
|
chr9_+_113433364
|
1.365
|
NM_001015882
NM_004125
|
DNAJC25
DNAJC25-GNG10
|
DnaJ (Hsp40) homolog, subfamily C , member 25
DNAJC25-GNG10 readthrough
|
|
chr3_-_191321601
|
1.364
|
NM_018192
|
LEPREL1
|
leprecan-like 1
|
|
chr13_-_71338558
|
1.360
|
|
DACH1
|
dachshund homolog 1 (Drosophila)
|
|
chr1_+_178390660
|
1.356
|
|
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1
|
|
chr14_-_51605515
|
1.348
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr2_+_20510312
|
1.343
|
NM_004040
|
RHOB
|
ras homolog gene family, member B
|
|
chr22_+_31527744
|
1.340
|
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr1_+_63832067
|
1.334
|
NM_001172819
|
PGM1
|
phosphoglucomutase 1
|
|
chr17_-_37221671
|
1.332
|
|
LEPREL4
|
leprecan-like 4
|
|
chr4_-_5071996
|
1.322
|
NM_018659
|
CYTL1
|
cytokine-like 1
|
|
chrX_-_92815222
|
1.321
|
NM_004538
|
NAP1L3
|
nucleosome assembly protein 1-like 3
|
|
chr14_-_93926679
|
1.320
|
NM_001002235
NM_001002236
NM_001127700
NM_001127701
NM_001127702
NM_001127703
NM_001127704
NM_001127705
NM_001127706
NM_001127707
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr9_-_14303899
|
1.296
|
|
NFIB
|
nuclear factor I/B
|
|
chr14_+_64076916
|
1.289
|
NM_021979
|
HSPA2
|
heat shock 70kDa protein 2
|
|
chr1_-_98159182
|
1.285
|
NM_000110
NM_001160301
|
DPYD
|
dihydropyrimidine dehydrogenase
|
|
chr12_-_55960501
|
1.274
|
|
R3HDM2
|
R3H domain containing 2
|
|
chr3_+_128190070
|
1.273
|
NM_032242
|
PLXNA1
|
plexin A1
|
|
chr14_-_51605695
|
1.268
|
NM_007361
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr5_+_52812165
|
1.265
|
|
FST
|
follistatin
|
|
chr14_+_89933055
|
1.259
|
NM_006888
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta)
|
|
chr16_-_87450849
|
1.257
|
NM_000512
|
GALNS
|
galactosamine (N-acetyl)-6-sulfate sulfatase
|
|
chr7_+_93862137
|
1.255
|
|
COL1A2
PLAG1
|
collagen, type I, alpha 2
pleiomorphic adenoma gene 1
|
|
chr1_-_223907283
|
1.250
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr13_+_34414390
|
1.244
|
NM_015678
|
NBEA
|
neurobeachin
|
|
chr14_-_74029788
|
1.231
|
NM_006432
|
NPC2
|
Niemann-Pick disease, type C2
|
|
chr16_+_55217085
|
1.226
|
NM_175617
|
MT1E
|
metallothionein 1E
|
|
chr14_-_74029720
|
1.223
|
|
NPC2
|
Niemann-Pick disease, type C2
|
|
chr15_+_72006015
|
1.222
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chrX_+_154264931
|
1.220
|
NM_001007523
NM_001007524
NM_012151
|
F8A2
F8A3
F8A1
|
coagulation factor VIII-associated (intronic transcript) 2
coagulation factor VIII-associated (intronic transcript) 3
coagulation factor VIII-associated (intronic transcript) 1
|
|
chr17_+_45488330
|
1.219
|
NM_002204
NM_005501
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr1_+_178390598
|
1.216
|
|
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1
|
|
chr11_-_11987115
|
1.215
|
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr11_-_65871592
|
1.213
|
NM_006876
|
B3GNT1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1
|
|
chr20_+_43953428
|
1.213
|
|
CTSA
|
cathepsin A
|
|
chr1_-_154913712
|
1.210
|
NM_006617
|
NES
|
nestin
|
|
chr3_-_151171190
|
1.209
|
NM_002628
NM_053024
|
PFN2
|
profilin 2
|
|
chr14_-_74029756
|
1.208
|
|
NPC2
|
Niemann-Pick disease, type C2
|
|
chr1_+_85819004
|
1.206
|
NM_001554
|
CYR61
|
cysteine-rich, angiogenic inducer, 61
|
|
chr15_-_46724360
|
1.197
|
|
FBN1
|
fibrillin 1
|
|
chr11_+_113671744
|
1.191
|
NM_006169
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr3_-_87122936
|
1.174
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr6_+_151688327
|
1.168
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr5_+_52812236
|
1.165
|
NM_006350
NM_013409
|
FST
|
follistatin
|
|
chr16_+_55217180
|
1.160
|
|
MT1E
|
metallothionein 1E
|
|
chr9_-_128924706
|
1.146
|
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr9_-_129656830
|
1.145
|
NM_000118
NM_001114753
|
ENG
|
endoglin
|
|
chr7_-_136678815
|
1.144
|
|
PTN
|
pleiotrophin
|
|
chr9_-_129656581
|
1.143
|
|
ENG
|
endoglin
|
|
chr11_-_102174102
|
1.142
|
NM_001145938
NM_002421
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase)
|
|
chr5_-_121440704
|
1.140
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr11_-_102174032
|
1.138
|
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase)
|
|
chr10_-_79067552
|
1.131
|
NM_001014797
NM_001161352
NM_001161353
NM_002247
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chr14_-_74491878
|
1.127
|
NM_002632
|
PGF
|
placental growth factor
|
|
chr1_-_98159113
|
1.118
|
|
DPYD
|
dihydropyrimidine dehydrogenase
|
|
chr1_+_51474558
|
1.116
|
|
RNF11
|
ring finger protein 11
|
|
chr19_+_50041400
|
1.115
|
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|