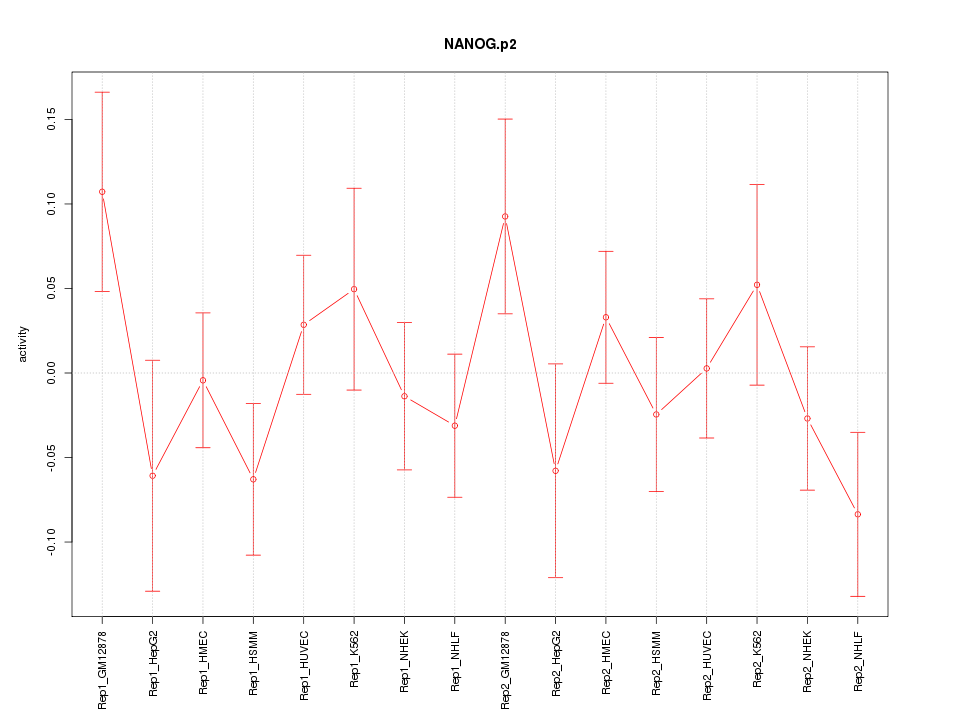
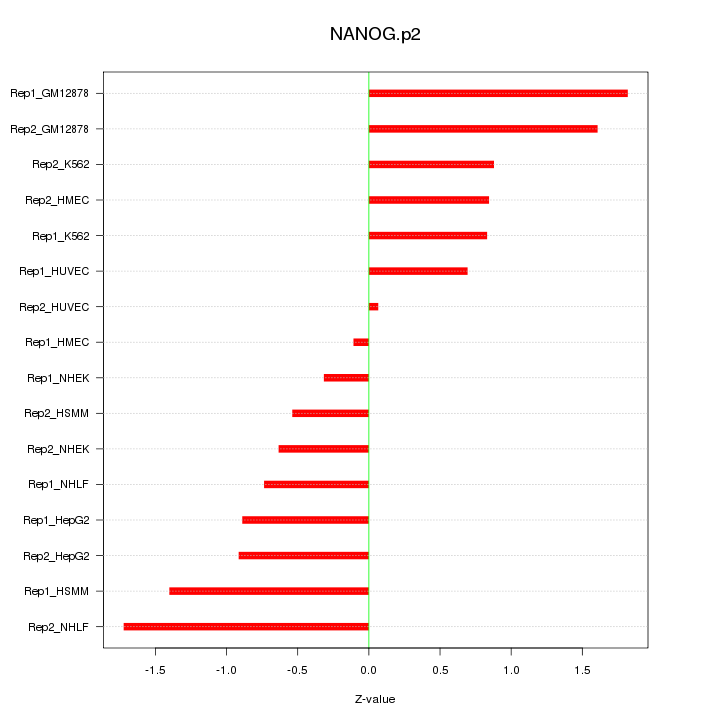
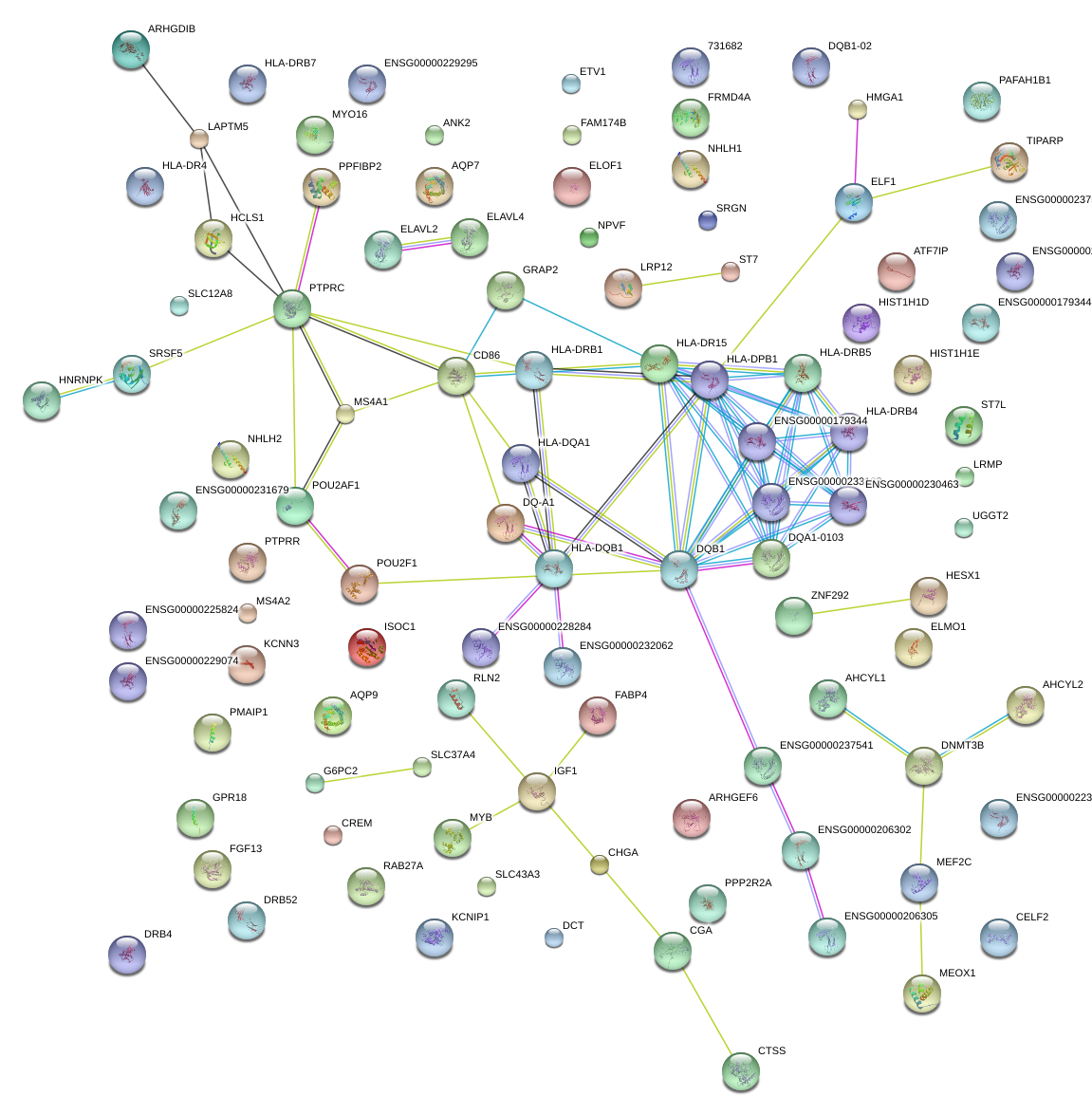
|
chr1_-_30977931
|
2.595
|
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chr12_-_15005761
|
2.549
|
NM_001175
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta
|
|
chr1_+_196874759
|
2.260
|
NM_002838
NM_080921
NM_080923
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr15_-_53349775
|
2.225
|
NM_183234
NM_004580
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr6_-_32605898
|
2.184
|
NM_002125
|
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 5
|
|
chr11_-_56951628
|
2.181
|
NM_017611
|
SLC43A3
|
solute carrier family 43, member 3
|
|
chr1_+_196874839
|
2.117
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr15_-_53350383
|
1.838
|
NM_183236
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr12_+_25096472
|
1.699
|
NM_006152
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr12_+_25096755
|
1.436
|
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr7_-_36992221
|
1.436
|
NM_130442
|
ELMO1
|
engulfment and cell motility 1
|
|
chr11_-_110755112
|
1.384
|
|
POU2AF1
|
POU class 2 associating factor 1
|
|
chr6_-_32665409
|
1.349
|
NM_002124
|
HLA-DRB1
HLA-DRB4
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 1
major histocompatibility complex, class II, DR beta 4
major histocompatibility complex, class II, DR beta 5
|
|
chr13_-_40491405
|
1.313
|
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr13_-_40491491
|
1.225
|
NM_172373
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr3_-_126325132
|
1.198
|
|
SLC12A8
|
solute carrier family 12 (potassium/chloride transporters), member 8
|
|
chr7_+_116447527
|
1.154
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr1_-_149004852
|
1.153
|
|
CTSS
|
cathepsin S
|
|
chr8_-_82557956
|
1.150
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr7_-_25234582
|
1.142
|
NM_022150
|
NPVF
|
neuropeptide VF precursor
|
|
chr10_+_11246934
|
1.091
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr6_-_26343139
|
1.090
|
NM_005320
|
HIST1H1D
|
histone cluster 1, H1d
|
|
chr11_+_59979857
|
1.049
|
NM_021950
NM_152866
|
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1
|
|
chr7_-_13992351
|
1.026
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr6_+_34314545
|
1.022
|
NM_145905
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr13_-_98708549
|
1.001
|
NM_001098200
NM_005292
|
GPR18
|
G protein-coupled receptor 18
|
|
chr12_-_101398451
|
0.994
|
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr3_-_57209319
|
0.947
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chr10_+_35524835
|
0.931
|
NM_182721
NM_182722
NM_182723
NM_182724
NM_182725
|
CREM
|
cAMP responsive element modulator
|
|
chr9_-_5294579
|
0.912
|
NM_005059
NM_134441
|
RLN2
|
relaxin 2
|
|
chr1_-_153109349
|
0.911
|
NM_002249
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr1_-_149004916
|
0.891
|
NM_004079
|
CTSS
|
cathepsin S
|
|
chr13_-_93929719
|
0.889
|
NM_001129889
NM_001922
|
DCT
|
dopachrome tautomerase (dopachrome delta-isomerase, tyrosine-related protein 2)
|
|
chr3_-_122862447
|
0.889
|
NM_005335
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr6_+_32713160
|
0.876
|
NM_002122
|
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1
|
|
chr3_-_122862391
|
0.869
|
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr6_+_34312906
|
0.868
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr12_+_14429499
|
0.865
|
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr13_+_108079570
|
0.842
|
NM_001198950
|
MYO16
|
myosin XVI
|
|
chr22_+_38652555
|
0.810
|
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr12_-_101398503
|
0.804
|
NM_000618
NM_001111283
NM_001111285
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr2_+_169465995
|
0.798
|
NM_001081686
NM_021176
|
G6PC2
|
glucose-6-phosphatase, catalytic, 2
|
|
chr8_+_26206648
|
0.789
|
NM_001177591
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr10_+_70517863
|
0.773
|
|
SRGN
|
serglycin
|
|
chr1_+_50342168
|
0.760
|
NM_001144776
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr6_+_135544214
|
0.734
|
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr1_+_165564904
|
0.729
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr12_-_101398429
|
0.724
|
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr10_-_14412813
|
0.711
|
NM_018027
|
FRMD4A
|
FERM domain containing 4A
|
|
chr1_+_165565006
|
0.709
|
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr1_+_50347188
|
0.707
|
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr3_+_123279439
|
0.706
|
NM_006889
|
CD86
|
CD86 molecule
|
|
chr5_-_88215034
|
0.697
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr6_+_88026338
|
0.683
|
|
ZNF292
|
zinc finger protein 292
|
|
chr17_-_1195096
|
0.681
|
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
|
|
chr6_+_34312979
|
0.676
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr11_-_118405288
|
0.674
|
NM_001164280
|
SLC37A4
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4
|
|
chr12_-_69434330
|
0.674
|
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr11_+_7574992
|
0.672
|
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr7_+_128802719
|
0.664
|
NM_001130722
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr5_+_169713458
|
0.650
|
NM_001034838
|
KCNIP1
|
Kv channel interacting protein 1
|
|
chr18_+_55718171
|
0.645
|
NM_021127
|
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1
|
|
chrX_-_137649180
|
0.642
|
NM_033642
|
FGF13
|
fibroblast growth factor 13
|
|
chr9_-_85773899
|
0.634
|
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K
|
|
chr12_-_101398470
|
0.631
|
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr1_-_116185215
|
0.629
|
NM_005599
|
NHLH2
|
nescient helix loop helix 2
|
|
chr20_+_30831298
|
0.619
|
NM_175850
|
DNMT3B
|
DNA (cytosine-5-)-methyltransferase 3 beta
|
|
chr10_-_13789628
|
0.618
|
|
FRMD4A
|
FERM domain containing 4A
|
|
chr17_-_39094787
|
0.615
|
NM_001040002
|
MEOX1
|
mesenchyme homeobox 1
|
|
chr14_+_69307914
|
0.610
|
|
SRSF5
|
serine/arginine-rich splicing factor 5
|
|
chr1_-_149004879
|
0.610
|
|
CTSS
|
cathepsin S
|
|
chr6_+_34312633
|
0.605
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr5_-_88235624
|
0.595
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr15_-_91078307
|
0.593
|
|
FAM174B
|
family with sequence similarity 174, member B
|
|
chr4_+_114433551
|
0.569
|
|
ANK2
|
ankyrin 2, neuronal
|
|
chr3_+_157877146
|
0.560
|
NM_001184718
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase
|
|
chr6_-_87861492
|
0.548
|
NM_000735
|
CGA
|
glycoprotein hormones, alpha polypeptide
|
|
chrX_-_135690699
|
0.547
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr5_+_128468785
|
0.545
|
|
ISOC1
|
isochorismatase domain containing 1
|
|
chr12_-_101398288
|
0.535
|
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr15_+_56217686
|
0.518
|
NM_020980
|
AQP9
|
aquaporin 9
|
|
chr1_+_196874867
|
0.516
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr4_-_123761615
|
0.515
|
NM_021803
|
IL21
|
interleukin 21
|
|
chr1_-_150653351
|
0.510
|
NM_016190
|
CRNN
|
cornulin
|
|
chr6_+_32515596
|
0.505
|
NM_019111
|
HLA-DRA
|
major histocompatibility complex, class II, DR alpha
|
|
chr11_+_75203859
|
0.503
|
NM_003369
|
UVRAG
|
UV radiation resistance associated gene
|
|
chr11_+_75203884
|
0.498
|
|
UVRAG
|
UV radiation resistance associated gene
|
|
chr12_+_52105131
|
0.496
|
|
AMHR2
|
anti-Mullerian hormone receptor, type II
|
|
chr5_-_169657395
|
0.492
|
NM_005565
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr9_+_12683385
|
0.491
|
NM_000550
|
TYRP1
|
tyrosinase-related protein 1
|
|
chr6_+_34312641
|
0.479
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr1_+_50347176
|
0.478
|
NM_001144774
NM_021952
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr20_+_2224612
|
0.465
|
NM_003245
|
TGM3
|
transglutaminase 3 (E polypeptide, protein-glutamine-gamma-glutamyltransferase)
|
|
chrX_+_119622570
|
0.463
|
NM_001137554
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr10_-_101680645
|
0.459
|
|
DNMBP
|
dynamin binding protein
|
|
chr12_-_101036407
|
0.458
|
NM_024057
|
NUP37
|
nucleoporin 37kDa
|
|
chr3_+_89239494
|
0.454
|
|
EPHA3
|
EPH receptor A3
|
|
chr12_-_23995109
|
0.449
|
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr4_+_146250978
|
0.449
|
|
ABCE1
|
ATP-binding cassette, sub-family E (OABP), member 1
|
|
chr6_+_34313007
|
0.447
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr6_-_154719560
|
0.439
|
NM_001130700
NM_015553
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1
|
|
chr2_+_161980865
|
0.439
|
NM_006593
|
TBR1
|
T-box, brain, 1
|
|
chr17_-_46479127
|
0.438
|
|
SPAG9
|
sperm associated antigen 9
|
|
chr1_+_154386358
|
0.435
|
NM_001193301
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr12_-_6103945
|
0.435
|
NM_000552
|
VWF
|
von Willebrand factor
|
|
chr16_+_55373241
|
0.429
|
|
NUP93
|
nucleoporin 93kDa
|
|
chr4_-_151990061
|
0.423
|
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing
|
|
chr3_+_195336625
|
0.422
|
NM_005524
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr3_+_88270951
|
0.421
|
NM_018293
|
ZNF654
|
zinc finger protein 654
|
|
chr4_-_176970895
|
0.418
|
|
GPM6A
|
glycoprotein M6A
|
|
chr8_-_40874485
|
0.417
|
NM_001135731
NM_024645
|
ZMAT4
|
zinc finger, matrin-type 4
|
|
chr2_+_161981046
|
0.416
|
|
TBR1
|
T-box, brain, 1
|
|
chrX_+_28515479
|
0.411
|
NM_014271
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1
|
|
chr6_+_34312625
|
0.398
|
NM_145901
NM_145902
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr5_+_137701122
|
0.398
|
NM_001135647
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr15_+_87432699
|
0.396
|
|
ABHD2
|
abhydrolase domain containing 2
|
|
chr6_-_32742265
|
0.390
|
NM_002123
|
HLA-DQB1
|
major histocompatibility complex, class II, DQ beta 1
|
|
chr2_+_161981138
|
0.380
|
|
TBR1
|
T-box, brain, 1
|
|
chr18_+_30812205
|
0.378
|
NM_001143827
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr8_-_102029997
|
0.378
|
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr3_+_154362726
|
0.377
|
|
RAP2B
|
RAP2B, member of RAS oncogene family
|
|
chr12_+_8167494
|
0.373
|
NM_016184
NM_194447
NM_194448
NM_194450
|
CLEC4A
|
C-type lectin domain family 4, member A
|
|
chr1_+_42934910
|
0.370
|
|
YBX1
|
Y box binding protein 1
|
|
chr18_-_30057166
|
0.368
|
|
NOL4
|
nucleolar protein 4
|
|
chr12_+_49155798
|
0.368
|
|
LARP4
|
La ribonucleoprotein domain family, member 4
|
|
chr14_+_100365162
|
0.365
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr6_+_26212082
|
0.363
|
NM_003542
|
HIST1H4C
|
histone cluster 1, H4c
|
|
chr11_+_58146721
|
0.363
|
NM_000614
|
CNTF
|
ciliary neurotrophic factor
|
|
chr6_-_55848346
|
0.362
|
|
BMP5
|
bone morphogenetic protein 5
|
|
chr20_+_42463419
|
0.362
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr15_-_68177283
|
0.358
|
NM_001105192
NM_005078
NM_020908
|
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila)
|
|
chr11_+_10433241
|
0.356
|
NM_001025389
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr11_-_33870411
|
0.355
|
NM_005574
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr15_+_87432735
|
0.355
|
|
ABHD2
|
abhydrolase domain containing 2
|
|
chr14_+_91859289
|
0.348
|
NM_153647
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr3_-_120861959
|
0.340
|
NM_022135
|
POPDC2
|
popeye domain containing 2
|
|
chr4_-_176971175
|
0.339
|
NM_201591
|
GPM6A
|
glycoprotein M6A
|
|
chr7_+_68702252
|
0.339
|
|
AUTS2
|
autism susceptibility candidate 2
|
|
chr1_-_198643885
|
0.336
|
|
ZNF281
|
zinc finger protein 281
|
|
chr4_-_41445693
|
0.335
|
NM_003924
|
PHOX2B
|
paired-like homeobox 2b
|
|
chr9_+_117955890
|
0.334
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr18_+_19826734
|
0.334
|
NM_153211
|
TTC39C
|
tetratricopeptide repeat domain 39C
|
|
chr20_+_42463387
|
0.331
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr6_-_112033752
|
0.329
|
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr13_+_48582389
|
0.329
|
NM_014923
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr3_+_63859114
|
0.325
|
NM_001177387
|
ATXN7
|
ataxin 7
|
|
chr14_+_91858677
|
0.319
|
NM_153648
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr6_-_161614962
|
0.318
|
NM_020133
|
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta)
|
|
chr1_-_42157077
|
0.318
|
NM_001127714
NM_024503
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3
|
|
chr2_-_40510923
|
0.316
|
NM_001112800
NM_001112801
NM_021097
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr15_+_87432629
|
0.315
|
|
ABHD2
|
abhydrolase domain containing 2
|
|
chr1_+_50348052
|
0.315
|
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr5_+_176744037
|
0.313
|
NM_001167579
NM_003052
|
SLC34A1
|
solute carrier family 34 (sodium phosphate), member 1
|
|
chr12_+_84792203
|
0.313
|
NM_006183
|
NTS
|
neurotensin
|
|
chr7_+_28692122
|
0.312
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr4_-_83566246
|
0.306
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr16_-_21221872
|
0.305
|
NM_001014444
NM_001888
|
CRYM
|
crystallin, mu
|
|
chr18_+_13601664
|
0.300
|
NM_001003674
NM_001003675
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr1_+_51208499
|
0.299
|
|
CDKN2C
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4)
|
|
chr14_+_31868229
|
0.294
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr22_+_21006814
|
0.290
|
|
|
|
|
chr12_-_107551783
|
0.288
|
NM_003006
|
SELPLG
|
selectin P ligand
|
|
chr13_+_30207668
|
0.288
|
NM_001629
|
ALOX5AP
|
arachidonate 5-lipoxygenase-activating protein
|
|
chr9_-_8723893
|
0.288
|
NM_001040712
NM_001171025
NM_130391
NM_130392
NM_130393
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr10_-_115603848
|
0.285
|
NM_014881
|
DCLRE1A
|
DNA cross-link repair 1A
|
|
chr11_+_88550687
|
0.284
|
NM_000372
|
TYR
|
tyrosinase (oculocutaneous albinism IA)
|
|
chr1_-_198644111
|
0.284
|
|
ZNF281
|
zinc finger protein 281
|
|
chr20_+_42463350
|
0.283
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr12_+_50981915
|
0.281
|
NM_002284
|
KRT86
|
keratin 86
|
|
chr17_-_9870292
|
0.279
|
NM_001130831
|
GAS7
|
growth arrest-specific 7
|
|
chr5_+_140790141
|
0.275
|
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr5_+_176786279
|
0.273
|
NM_001004105
NM_001004106
NM_002082
|
GRK6
|
G protein-coupled receptor kinase 6
|
|
chr4_-_26100986
|
0.273
|
NM_000730
|
CCKAR
|
cholecystokinin A receptor
|
|
chr2_+_128119908
|
0.271
|
NM_001161415
NM_001161416
NM_001161417
NM_005291
|
GPR17
|
G protein-coupled receptor 17
|
|
chr7_+_50318801
|
0.270
|
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr5_-_169657312
|
0.269
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr16_+_55950195
|
0.269
|
NM_002990
|
CCL22
|
chemokine (C-C motif) ligand 22
|
|
chr20_+_42463279
|
0.266
|
NM_000457
NM_178849
NM_178850
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr6_+_49575629
|
0.265
|
NM_001010904
|
GLYATL3
|
glycine-N-acyltransferase-like 3
|
|
chr8_-_102872372
|
0.264
|
|
NCALD
|
neurocalcin delta
|
|
chr11_-_102001259
|
0.262
|
NM_004771
|
MMP20
|
matrix metallopeptidase 20
|
|
chr12_-_52281050
|
0.262
|
NM_001130059
|
ATF7
|
activating transcription factor 7
|
|
chr7_+_150321771
|
0.259
|
NM_001160109
NM_001160110
NM_001160111
|
NOS3
|
nitric oxide synthase 3 (endothelial cell)
|
|
chr10_+_35456095
|
0.258
|
|
CREM
|
cAMP responsive element modulator
|
|
chr11_+_20577521
|
0.256
|
NM_004211
|
SLC6A5
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 5
|
|
chr7_+_18501876
|
0.253
|
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chr10_+_35456393
|
0.253
|
NM_181571
NM_183060
|
CREM
|
cAMP responsive element modulator
|
|
chr3_-_102522093
|
0.252
|
NM_016247
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2
|
|
chr14_-_22496098
|
0.252
|
NM_001166269
NM_001166270
NM_017815
|
HAUS4
|
HAUS augmin-like complex, subunit 4
|
|
chr9_+_115265827
|
0.252
|
NM_017790
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr7_+_115650240
|
0.251
|
NM_152829
|
TES
|
testis derived transcript (3 LIM domains)
|
|
chr6_-_24827265
|
0.249
|
NM_030939
|
C6orf62
|
chromosome 6 open reading frame 62
|
|
chr3_+_138132006
|
0.248
|
NM_001190796
|
NCK1
|
NCK adaptor protein 1
|
|
chr20_+_36365965
|
0.246
|
NM_001725
|
BPI
|
bactericidal/permeability-increasing protein
|
|
chr6_+_16251260
|
0.245
|
|
MYLIP
|
myosin regulatory light chain interacting protein
|
|
chr6_-_112034166
|
0.242
|
NM_001164281
NM_147686
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr1_+_74473672
|
0.242
|
NM_015978
|
TNNI3K
|
TNNI3 interacting kinase
|
|
chrX_-_123925343
|
0.242
|
NM_001163278
NM_001163279
NM_014253
|
ODZ1
|
odz, odd Oz/ten-m homolog 1(Drosophila)
|
|
chr18_-_30057512
|
0.240
|
NM_001198546
NM_001198548
NM_003787
|
NOL4
|
nucleolar protein 4
|
|
chr6_-_55848317
|
0.239
|
NM_021073
|
BMP5
|
bone morphogenetic protein 5
|