|
chr17_-_35975227
|
6.134
|
NM_001838
|
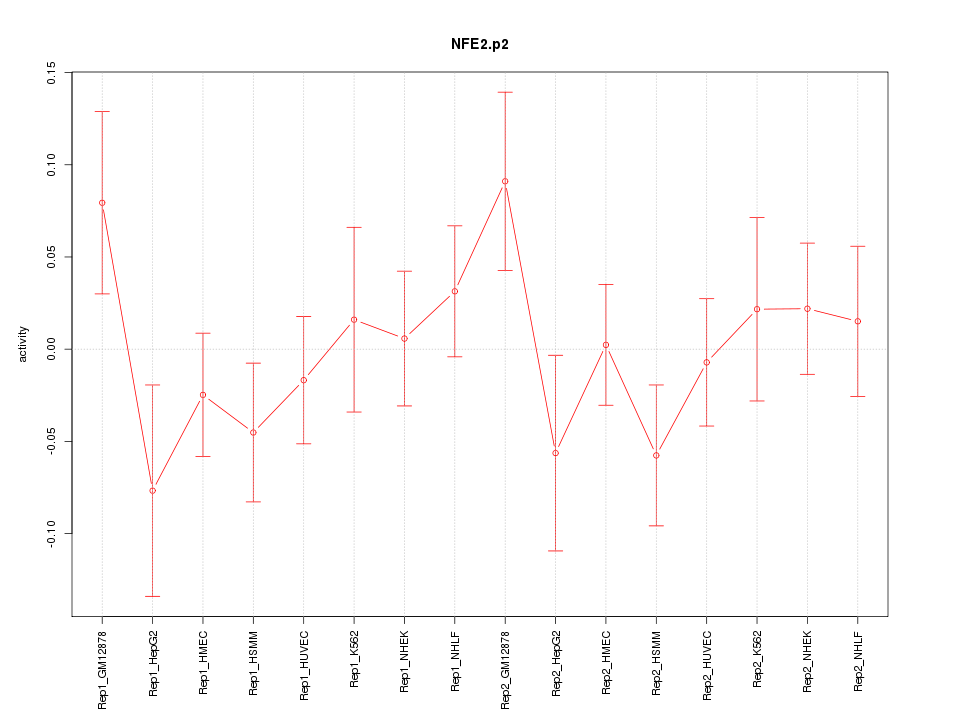
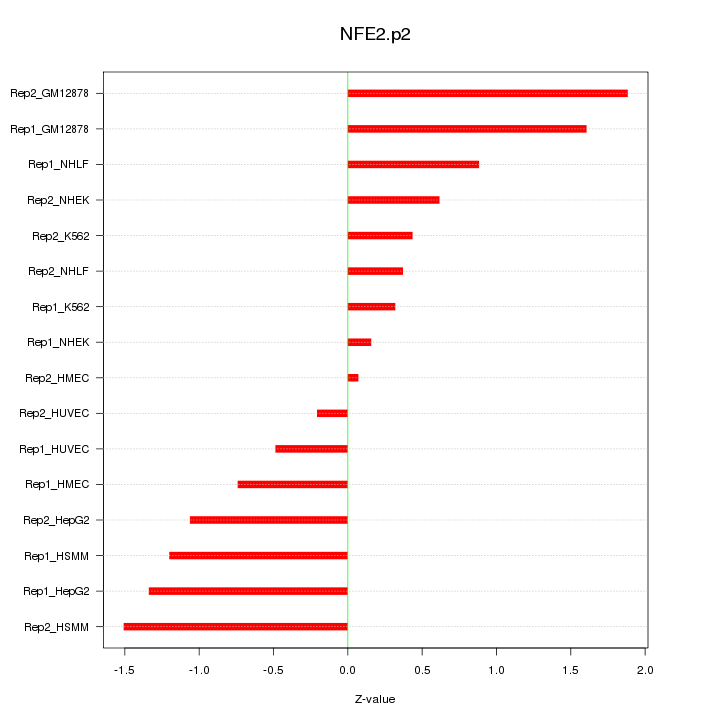
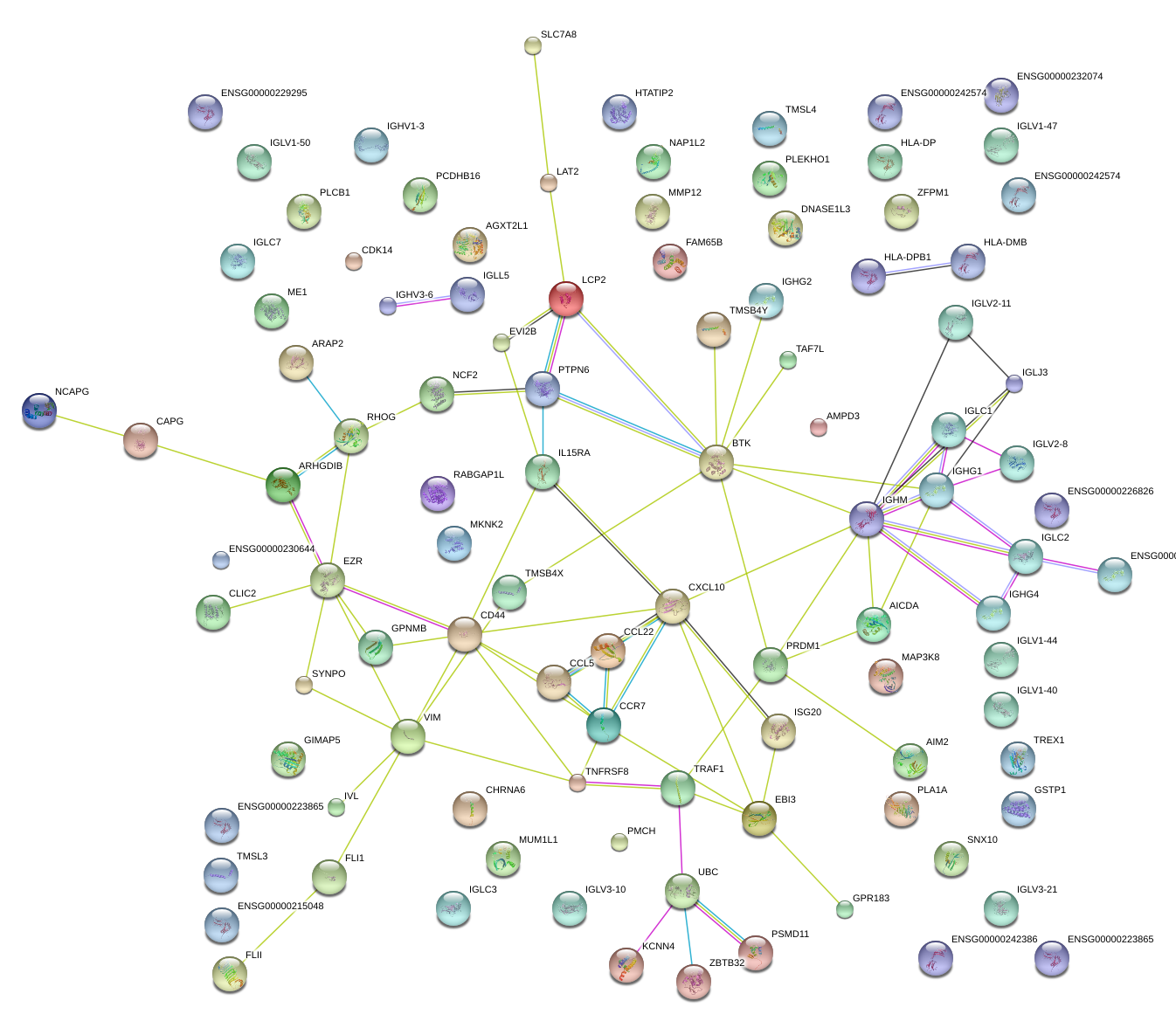
CCR7
|
chemokine (C-C motif) receptor 7
|
|
chr22_+_21495273
|
5.294
|
|
|
|
|
chr22_+_21370394
|
5.090
|
|
IGL@
|
immunoglobulin lambda locus
|
|
chr22_+_21053972
|
4.104
|
|
|
|
|
chr22_+_21011653
|
3.020
|
|
|
|
|
chr22_+_21116283
|
3.012
|
|
|
|
|
chr12_-_15005761
|
2.945
|
NM_001175
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta
|
|
chr1_+_173111306
|
2.837
|
NM_001035230
|
RABGAP1L
|
RAB GTPase activating protein 1-like
|
|
chr13_-_98757634
|
2.825
|
NM_004951
|
GPR183
|
G protein-coupled receptor 183
|
|
chr22_+_21340757
|
2.687
|
|
|
|
|
chr22_+_20899155
|
2.600
|
|
|
|
|
chr22_+_21065134
|
2.411
|
|
LOC100290481
|
immunoglobulin lambda light chain-like
|
|
chr22_+_21079355
|
2.407
|
|
|
|
|
chr17_-_26665144
|
2.373
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr17_-_26665213
|
2.325
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chrX_-_154216929
|
2.272
|
NM_001289
|
CLIC2
|
chloride intracellular channel 2
|
|
chr22_+_21065207
|
2.206
|
|
IGLJ3
|
immunoglobulin lambda joining 3
|
|
chr17_-_26665194
|
2.176
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr6_+_33151703
|
2.116
|
NM_002121
|
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1
|
|
chr22_+_21464980
|
1.989
|
|
|
|
|
chr11_+_67107641
|
1.926
|
NM_000852
|
GSTP1
|
glutathione S-transferase pi 1
|
|
chr11_+_67107852
|
1.903
|
|
GSTP1
|
glutathione S-transferase pi 1
|
|
chr9_-_122728173
|
1.858
|
|
TRAF1
|
TNF receptor-associated factor 1
|
|
chrX_+_12903145
|
1.855
|
NM_021109
NM_183049
|
TMSB4X
TMSL3
|
thymosin beta 4, X-linked
thymosin-like 3
|
|
chr11_+_20341806
|
1.800
|
NM_006410
NM_001098521
|
HTATIP2
|
HIV-1 Tat interactive protein 2, 30kDa
|
|
chr11_+_20341900
|
1.740
|
NM_001098522
|
HTATIP2
|
HIV-1 Tat interactive protein 2, 30kDa
|
|
chrX_-_100527799
|
1.735
|
NM_000061
|
BTK
|
Bruton agammaglobulinemia tyrosine kinase
|
|
chr11_+_20341835
|
1.658
|
NM_001098520
|
HTATIP2
|
HIV-1 Tat interactive protein 2, 30kDa
|
|
chr17_-_26665220
|
1.605
|
NM_006495
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr6_-_84197477
|
1.556
|
|
ME1
|
malic enzyme 1, NADP(+)-dependent, cytosolic
|
|
chr22_+_21552897
|
1.521
|
|
LOC100290481
|
immunoglobulin lambda light chain-like
|
|
chr6_-_84197370
|
1.515
|
|
ME1
|
malic enzyme 1, NADP(+)-dependent, cytosolic
|
|
chr15_+_86983038
|
1.470
|
NM_002201
|
ISG20
|
interferon stimulated exonuclease gene 20kDa
|
|
chr22_+_21042091
|
1.411
|
|
IGLV1-44
IGL@
|
immunoglobulin lambda variable 1-44
immunoglobulin lambda locus
|
|
chr6_-_84197463
|
1.384
|
|
ME1
|
malic enzyme 1, NADP(+)-dependent, cytosolic
|
|
chr14_-_105280425
|
1.329
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr4_-_77163609
|
1.308
|
NM_001565
|
CXCL10
|
chemokine (C-X-C motif) ligand 10
|
|
chr12_+_6930694
|
1.267
|
NM_002831
NM_080549
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr1_+_12046020
|
1.162
|
NM_001243
|
TNFRSF8
|
tumor necrosis factor receptor superfamily, member 8
|
|
chr19_+_4180494
|
1.143
|
NM_005755
|
EBI3
|
Epstein-Barr virus induced 3
|
|
chr7_+_23252915
|
1.137
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr5_+_150000394
|
1.126
|
NM_001109974
NM_007286
|
SYNPO
|
synaptopodin
|
|
chr11_+_20342262
|
1.124
|
NM_001098523
|
HTATIP2
|
HIV-1 Tat interactive protein 2, 30kDa
|
|
chr7_+_23252802
|
1.120
|
NM_001005340
NM_002510
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr7_+_73262283
|
1.112
|
|
LAT2
|
linker for activation of T cells family, member 2
|
|
chr5_-_169657312
|
1.111
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr14_-_105308720
|
1.110
|
|
IGHM
|
immunoglobulin heavy constant mu
|
|
chr6_-_84197573
|
1.096
|
NM_002395
|
ME1
|
malic enzyme 1, NADP(+)-dependent, cytosolic
|
|
chr14_-_105308639
|
1.093
|
|
|
|
|
chr1_-_181826292
|
1.069
|
NM_000433
NM_001190789
NM_001190794
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr10_+_30763056
|
1.063
|
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8
|
|
chr5_-_169657395
|
1.042
|
NM_005565
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr10_-_6059460
|
1.041
|
NM_002189
NM_172200
|
IL15RA
|
interleukin 15 receptor, alpha
|
|
chrX_-_72351381
|
1.039
|
|
NAP1L2
|
nucleosome assembly protein 1-like 2
|
|
chr22_+_21065202
|
1.034
|
|
|
|
|
chr22_+_21006814
|
1.032
|
|
|
|
|
chr7_+_73262264
|
1.024
|
|
LAT2
|
linker for activation of T cells family, member 2
|
|
chr7_+_23252911
|
1.015
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr3_+_48482213
|
1.013
|
NM_016381
NM_033629
|
TREX1
|
three prime repair exonuclease 1
|
|
chr4_-_35922288
|
1.007
|
NM_015230
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2
|
|
chrX_-_72351348
|
0.993
|
|
NAP1L2
|
nucleosome assembly protein 1-like 2
|
|
chr19_+_40895669
|
0.987
|
NM_014383
|
ZBTB32
|
zinc finger and BTB domain containing 32
|
|
chr10_+_30762862
|
0.972
|
NM_005204
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8
|
|
chr14_-_105163301
|
0.965
|
|
IGHG4
|
immunoglobulin heavy constant gamma 4 (G4m marker)
|
|
chr1_+_148388870
|
0.945
|
|
PLEKHO1
|
pleckstrin homology domain containing, family O member 1
|
|
chrX_-_72351392
|
0.943
|
NM_021963
|
NAP1L2
|
nucleosome assembly protein 1-like 2
|
|
chr17_+_27795633
|
0.937
|
|
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11
|
|
chr3_-_58175437
|
0.929
|
NM_004944
|
DNASE1L3
|
deoxyribonuclease I-like 3
|
|
chr7_+_90177082
|
0.928
|
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr8_-_42742729
|
0.920
|
NM_004198
|
CHRNA6
|
cholinergic receptor, nicotinic, alpha 6
|
|
chr3_+_48482625
|
0.905
|
|
TREX1
|
three prime repair exonuclease 1
|
|
chr11_+_10433241
|
0.889
|
NM_001025389
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr5_-_169657360
|
0.886
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr6_-_159159242
|
0.883
|
|
EZR
|
ezrin
|
|
chr3_+_120799411
|
0.875
|
NM_015900
|
PLA1A
|
phospholipase A1 member A
|
|
chr10_+_17310263
|
0.869
|
NM_003380
|
VIM
|
vimentin
|
|
chr7_+_23252945
|
0.866
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr10_+_17310475
|
0.857
|
|
VIM
|
vimentin
|
|
chr16_+_55950195
|
0.841
|
NM_002990
|
CCL22
|
chemokine (C-C motif) ligand 22
|
|
chr6_-_159159186
|
0.827
|
|
EZR
|
ezrin
|
|
chr1_+_148389039
|
0.824
|
|
PLEKHO1
|
pleckstrin homology domain containing, family O member 1
|
|
chr1_+_148388719
|
0.824
|
NM_016274
|
PLEKHO1
|
pleckstrin homology domain containing, family O member 1
|
|
chr11_+_35117226
|
0.821
|
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr1_-_157313179
|
0.820
|
NM_004833
|
AIM2
|
absent in melanoma 2
|
|
chr14_-_105182025
|
0.810
|
|
|
|
|
chr6_-_159159257
|
0.808
|
NM_003379
|
EZR
|
ezrin
|
|
chr1_+_151147646
|
0.805
|
NM_005547
|
IVL
|
involucrin
|
|
chr1_-_181826086
|
0.787
|
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr12_-_8656705
|
0.787
|
NM_020661
|
AICDA
|
activation-induced cytidine deaminase
|
|
chr22_+_21027538
|
0.769
|
|
|
|
|
chr7_+_23252909
|
0.767
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr7_+_26298018
|
0.755
|
NM_013322
|
SNX10
|
sorting nexin 10
|
|
chr17_-_31231421
|
0.747
|
NM_002985
|
CCL5
|
chemokine (C-C motif) ligand 5
|
|
chr12_-_101115677
|
0.745
|
NM_002674
|
PMCH
|
pro-melanin-concentrating hormone
|
|
chr11_-_3818713
|
0.731
|
|
RHOG
|
ras homolog gene family, member G (rho G)
|
|
chr7_+_26298136
|
0.730
|
|
SNX10
|
sorting nexin 10
|
|
chr11_-_3818759
|
0.710
|
NM_001665
|
RHOG
|
ras homolog gene family, member G (rho G)
|
|
chr19_-_1989937
|
0.702
|
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chr6_+_106653429
|
0.697
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr19_-_48976852
|
0.691
|
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr7_+_150065376
|
0.683
|
NM_018384
|
GIMAP5
|
GTPase, IMAP family member 5
|
|
chr6_-_24985432
|
0.680
|
NM_015864
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chr2_-_85494639
|
0.675
|
|
CAPG
|
capping protein (actin filament), gelsolin-like
|
|
chr6_-_33016561
|
0.673
|
NM_002118
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta
|
|
chr11_+_128068982
|
0.661
|
NM_002017
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr3_-_99724047
|
0.657
|
|
CLDND1
|
claudin domain containing 1
|
|
chr2_-_85494664
|
0.657
|
|
CAPG
|
capping protein (actin filament), gelsolin-like
|
|
chr8_-_70908130
|
0.652
|
NM_001146008
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1
|
|
chr6_-_31658044
|
0.645
|
NM_002341
NM_009588
|
LTB
|
lymphotoxin beta (TNF superfamily, member 3)
|
|
chr1_+_181421984
|
0.643
|
|
LAMC2
|
laminin, gamma 2
|
|
chr22_+_20880159
|
0.617
|
|
IGLV6-57
|
immunoglobulin lambda variable 6-57
|
|
chr11_+_10434056
|
0.614
|
NM_001025390
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr5_-_169657192
|
0.612
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr17_-_43254132
|
0.604
|
NM_145798
|
OSBPL7
|
oxysterol binding protein-like 7
|
|
chr16_-_87570716
|
0.601
|
NM_005187
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr4_+_170817787
|
0.590
|
|
CLCN3
|
chloride channel 3
|
|
chr11_-_58099884
|
0.586
|
NM_004811
|
LPXN
|
leupaxin
|
|
chr22_+_20880112
|
0.581
|
|
|
|
|
chr2_+_151922350
|
0.580
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr6_+_84626113
|
0.569
|
|
CYB5R4
|
cytochrome b5 reductase 4
|
|
chr11_+_35154775
|
0.569
|
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr19_-_48977247
|
0.568
|
NM_002250
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr11_-_62231242
|
0.565
|
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr11_-_110755112
|
0.561
|
|
POU2AF1
|
POU class 2 associating factor 1
|
|
chr17_-_25642962
|
0.561
|
NM_000386
|
BLMH
|
bleomycin hydrolase
|
|
chr3_-_99724373
|
0.560
|
|
CLDND1
|
claudin domain containing 1
|
|
chr8_-_70907957
|
0.556
|
NM_001146009
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1
|
|
chr17_-_43254117
|
0.550
|
|
OSBPL7
|
oxysterol binding protein-like 7
|
|
chr7_+_139175420
|
0.544
|
NM_001061
NM_001166253
NM_030984
|
TBXAS1
|
thromboxane A synthase 1 (platelet)
|
|
chr9_+_4975082
|
0.543
|
|
JAK2
|
Janus kinase 2
|
|
chr14_-_105182159
|
0.542
|
|
IGHG2
|
immunoglobulin heavy constant gamma 2 (G2m marker)
|
|
chr5_-_137099361
|
0.541
|
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chr11_+_128069203
|
0.540
|
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr18_-_51219880
|
0.533
|
|
TCF4
|
transcription factor 4
|
|
chr7_+_150065594
|
0.530
|
|
GIMAP5
|
GTPase, IMAP family member 5
|
|
chr6_+_84626146
|
0.527
|
|
CYB5R4
|
cytochrome b5 reductase 4
|
|
chr6_+_107066422
|
0.520
|
NM_001624
|
AIM1
|
absent in melanoma 1
|
|
chr6_+_84626083
|
0.517
|
NM_016230
|
CYB5R4
|
cytochrome b5 reductase 4
|
|
chr3_+_142525744
|
0.514
|
NM_001080412
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr2_-_175170674
|
0.513
|
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr3_-_99724438
|
0.513
|
|
CLDND1
|
claudin domain containing 1
|
|
chr5_-_137099602
|
0.511
|
NM_017415
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chr10_+_70745615
|
0.511
|
NM_033496
|
HK1
|
hexokinase 1
|
|
chr19_-_61039939
|
0.502
|
NM_145007
|
NLRP11
|
NLR family, pyrin domain containing 11
|
|
chr10_-_94323808
|
0.501
|
|
IDE
|
insulin-degrading enzyme
|
|
chr9_+_2005218
|
0.500
|
NM_003070
NM_139045
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2
|
|
chrX_-_129072152
|
0.499
|
NM_001421
|
ELF4
|
E74-like factor 4 (ets domain transcription factor)
|
|
chr11_+_35117242
|
0.497
|
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr2_-_89400935
|
0.494
|
|
IGKC
|
immunoglobulin kappa constant
|
|
chr11_-_102174102
|
0.491
|
NM_001145938
NM_002421
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase)
|
|
chr10_-_94323701
|
0.490
|
|
IDE
|
insulin-degrading enzyme
|
|
chr1_+_13948448
|
0.489
|
NM_001007257
|
PRDM2
|
PR domain containing 2, with ZNF domain
|
|
chr22_-_30388165
|
0.487
|
|
PISD
|
phosphatidylserine decarboxylase
|
|
chr15_-_73818013
|
0.485
|
|
|
|
|
chr4_-_87500201
|
0.482
|
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr4_-_151990061
|
0.480
|
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing
|
|
chr11_-_62231445
|
0.478
|
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr1_+_181421796
|
0.475
|
NM_005562
NM_018891
|
LAMC2
|
laminin, gamma 2
|
|
chr7_+_90176647
|
0.455
|
NM_012395
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr3_-_99724412
|
0.452
|
|
CLDND1
|
claudin domain containing 1
|
|
chr7_+_68701704
|
0.446
|
NM_001127231
NM_001127232
NM_015570
|
AUTS2
|
autism susceptibility candidate 2
|
|
chr1_-_146543016
|
0.445
|
|
|
|
|
chr18_+_59705918
|
0.440
|
NM_001143818
NM_002575
|
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2
|
|
chr1_+_40508336
|
0.439
|
|
ZMPSTE24
|
zinc metallopeptidase (STE24 homolog, S. cerevisiae)
|
|
chr22_+_20715340
|
0.438
|
|
|
|
|
chr17_-_70662364
|
0.431
|
NM_001002032
NM_001002033
NM_016185
|
HN1
|
hematological and neurological expressed 1
|
|
chr10_-_101759630
|
0.430
|
NM_015221
|
DNMBP
|
dynamin binding protein
|
|
chr22_+_21094094
|
0.429
|
|
|
|
|
chr9_+_4975243
|
0.425
|
NM_004972
|
JAK2
|
Janus kinase 2
|
|
chr4_+_40953698
|
0.422
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr22_-_37858871
|
0.420
|
|
CBX7
|
chromobox homolog 7
|
|
chr3_+_49002285
|
0.419
|
NM_177938
NM_177939
|
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum)
|
|
chr11_-_102174032
|
0.412
|
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase)
|
|
chr22_-_35875890
|
0.411
|
NM_000878
|
IL2RB
|
interleukin 2 receptor, beta
|
|
chr19_-_6542012
|
0.407
|
|
CD70
|
CD70 molecule
|
|
chr15_+_99276982
|
0.406
|
NM_024652
|
LRRK1
|
leucine-rich repeat kinase 1
|
|
chr1_-_223683141
|
0.405
|
NM_194442
|
LBR
|
lamin B receptor
|
|
chrX_-_16640590
|
0.405
|
NM_175859
NM_019857
|
CTPS2
|
CTP synthase II
|
|
chr19_-_40939756
|
0.405
|
NM_144617
|
HSPB6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chr17_+_69974116
|
0.403
|
NM_007261
|
CD300A
|
CD300a molecule
|
|
chr19_+_59820413
|
0.398
|
NM_006669
|
LILRB1
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 1
|
|
chr12_+_67489063
|
0.397
|
|
MDM2
|
Mdm2 p53 binding protein homolog (mouse)
|
|
chr11_+_118463906
|
0.394
|
NM_001024382
|
HMBS
|
hydroxymethylbilane synthase
|
|
chr3_+_133855979
|
0.393
|
NM_198329
|
UBA5
|
ubiquitin-like modifier activating enzyme 5
|
|
chr11_-_102219450
|
0.390
|
NM_002422
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase)
|
|
chr11_+_35116961
|
0.388
|
NM_000610
NM_001001389
NM_001001390
NM_001001391
NM_001001392
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr5_-_142795269
|
0.387
|
NM_001018074
NM_001018075
NM_001018077
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr20_+_44180355
|
0.380
|
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5
|
|
chr19_+_6482009
|
0.376
|
NM_003811
|
TNFSF9
|
tumor necrosis factor (ligand) superfamily, member 9
|
|
chr17_-_37022465
|
0.376
|
NM_005557
|
KRT16
|
keratin 16
|
|
chrX_-_129072103
|
0.373
|
|
ELF4
|
E74-like factor 4 (ets domain transcription factor)
|
|
chr10_-_94323827
|
0.373
|
NM_004969
|
IDE
|
insulin-degrading enzyme
|
|
chr4_-_38710376
|
0.371
|
NM_024943
|
TMEM156
|
transmembrane protein 156
|
|
chr19_-_6621078
|
0.371
|
|
TNFSF14
|
tumor necrosis factor (ligand) superfamily, member 14
|
|
chr9_-_73715666
|
0.367
|
|
FAM108B1
|
family with sequence similarity 108, member B1
|
|
chr9_-_34652629
|
0.364
|
NM_006664
|
CCL27
|
chemokine (C-C motif) ligand 27
|
|
chr19_+_43971689
|
0.361
|
NM_001042507
|
LGALS7B
|
lectin, galactoside-binding, soluble, 7B
|
|
chrX_-_47374187
|
0.361
|
NM_001145252
|
CFP
|
complement factor properdin
|
|
chr1_-_156923111
|
0.360
|
NM_003126
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2)
|
|
chr20_+_44180296
|
0.360
|
NM_001250
NM_152854
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5
|