|
chr1_-_150233337
|
12.113
|
NM_002966
|
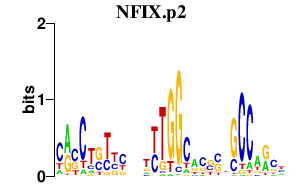
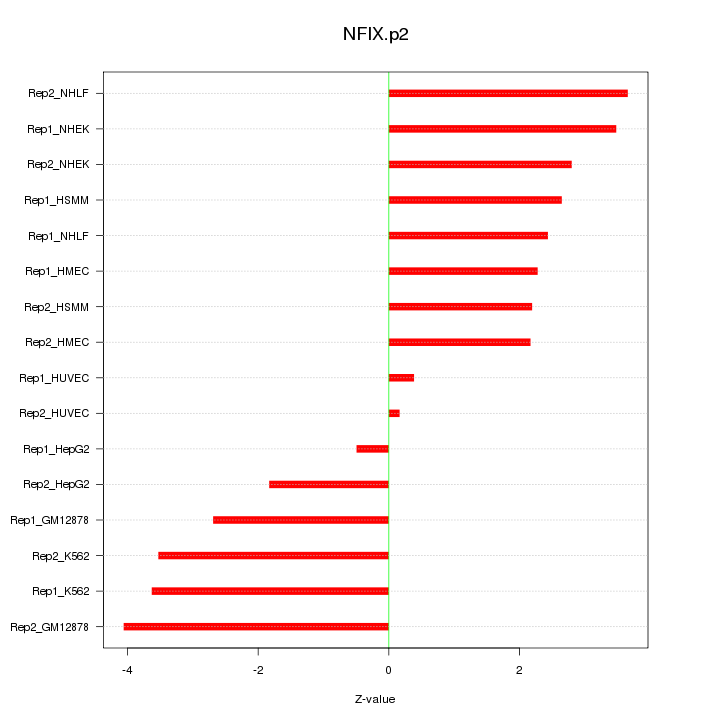
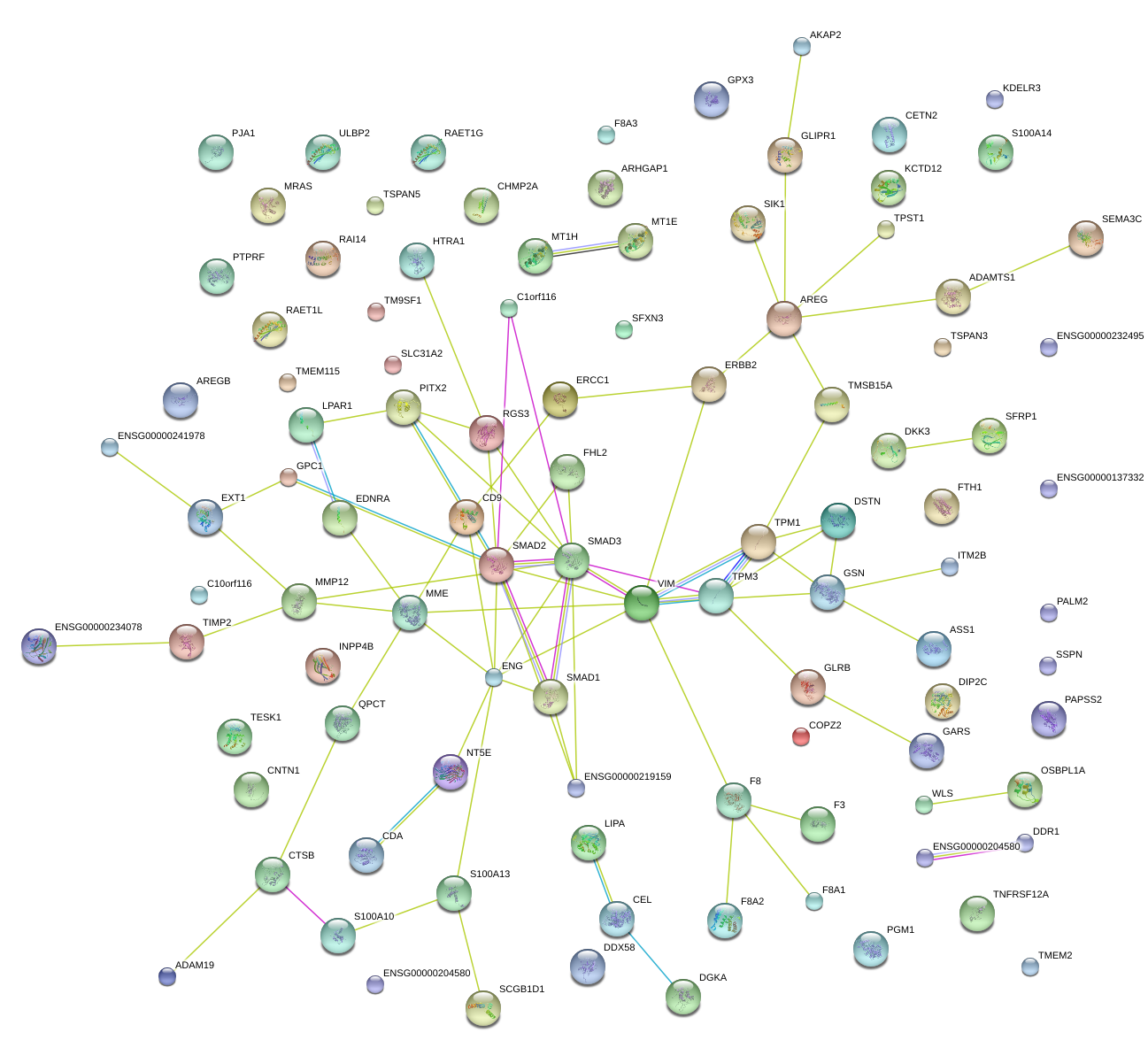
S100A10
|
S100 calcium binding protein A10
|
|
chr1_-_150232927
|
11.577
|
|
S100A10
|
S100 calcium binding protein A10
|
|
chr1_-_150232900
|
11.245
|
|
S100A10
|
S100 calcium binding protein A10
|
|
chr10_+_124210980
|
8.077
|
NM_002775
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr10_+_88718167
|
6.493
|
NM_006829
|
C10orf116
|
chromosome 10 open reading frame 116
|
|
chr15_+_61123327
|
6.373
|
|
TPM1
|
tropomyosin 1 (alpha)
|
|
chr16_+_3010311
|
5.751
|
NM_016639
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A
|
|
chr10_+_17312654
|
5.407
|
|
VIM
|
vimentin
|
|
chr10_+_17311788
|
5.316
|
|
VIM
|
vimentin
|
|
chr7_-_80386602
|
5.308
|
NM_006379
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C
|
|
chr10_+_17311962
|
5.123
|
|
VIM
|
vimentin
|
|
chr4_-_99798563
|
5.026
|
NM_005723
|
TSPAN5
|
tetraspanin 5
|
|
chr7_+_65307816
|
4.989
|
|
TPST1
|
tyrosylprotein sulfotransferase 1
|
|
chr7_-_80386258
|
4.542
|
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C
|
|
chr8_-_41286088
|
4.384
|
NM_003012
|
SFRP1
|
secreted frizzled-related protein 1
|
|
chr7_+_65307691
|
4.270
|
NM_003596
|
TPST1
|
tyrosylprotein sulfotransferase 1
|
|
chr9_+_123070191
|
4.042
|
NM_001127662
NM_001127663
NM_001127664
NM_001127665
NM_001127666
NM_001127667
NM_198252
|
GSN
|
gelsolin
|
|
chr2_+_241023703
|
3.958
|
NM_002081
|
GPC1
|
glypican 1
|
|
chr9_+_123070276
|
3.935
|
|
GSN
|
gelsolin
|
|
chr10_+_102782151
|
3.893
|
|
SFXN3
|
sideroflexin 3
|
|
chr9_+_123070234
|
3.795
|
|
GSN
|
gelsolin
|
|
chr10_+_124211353
|
3.683
|
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr11_-_11986704
|
3.575
|
NM_001018057
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr10_-_91001496
|
3.504
|
|
LIPA
|
lipase A, lysosomal acid, cholesterol esterase
|
|
chr21_-_27139563
|
3.477
|
NM_006988
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr9_+_114953048
|
3.330
|
NM_001860
|
SLC31A2
|
solute carrier family 31 (copper transporters), member 2
|
|
chr10_-_91001566
|
3.183
|
NM_000235
NM_001127605
|
LIPA
|
lipase A, lysosomal acid, cholesterol esterase
|
|
chr10_-_91001531
|
3.182
|
|
LIPA
|
lipase A, lysosomal acid, cholesterol esterase
|
|
chr1_+_43769065
|
3.181
|
NM_002840
NM_130440
|
PTPRF
|
protein tyrosine phosphatase, receptor type, F
|
|
chr10_-_91001542
|
3.073
|
|
LIPA
|
lipase A, lysosomal acid, cholesterol esterase
|
|
chr12_+_26239535
|
3.070
|
NM_001135823
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr1_+_63832067
|
3.003
|
NM_001172819
|
PGM1
|
phosphoglucomutase 1
|
|
chr5_+_34692239
|
2.962
|
|
RAI14
|
retinoic acid induced 14
|
|
chrX_-_101658297
|
2.931
|
NM_021992
|
TMSB15A
TMSB15B
|
thymosin beta 15a
thymosin beta 15B
|
|
chr17_-_74432800
|
2.790
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr13_-_76358433
|
2.718
|
NM_138444
|
KCTD12
|
potassium channel tetramerisation domain containing 12
|
|
chr3_+_156280129
|
2.715
|
NM_000902
|
MME
|
membrane metallo-endopeptidase
|
|
chr12_+_6179736
|
2.683
|
NM_001769
|
CD9
|
CD9 molecule
|
|
chr4_-_143987053
|
2.674
|
NM_001101669
NM_003866
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa
|
|
chr5_+_34692352
|
2.666
|
NM_001145520
|
RAI14
|
retinoic acid induced 14
|
|
chr5_+_34692124
|
2.564
|
NM_001145522
NM_015577
|
RAI14
|
retinoic acid induced 14
|
|
chr9_+_132309914
|
2.557
|
NM_000050
NM_054012
|
ASS1
|
argininosuccinate synthase 1
|
|
chr6_+_86216020
|
2.546
|
NM_002526
|
NT5E
|
5'-nucleotidase, ecto (CD73)
|
|
chr1_+_20788027
|
2.510
|
NM_001785
|
CDA
|
cytidine deaminase
|
|
chr12_+_26239755
|
2.444
|
NM_005086
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr16_+_55261226
|
2.372
|
NM_005951
|
MT1H
|
metallothionein 1H
|
|
chr21_-_27139143
|
2.343
|
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr9_+_111582317
|
2.284
|
NM_053016
NM_007203
NM_147150
|
PALM2
PALM2-AKAP2
|
paralemmin 2
PALM2-AKAP2 readthrough
|
|
chr11_-_61491684
|
2.228
|
NM_002032
|
FTH1
|
ferritin, heavy polypeptide 1
|
|
chr9_+_111850698
|
2.213
|
NM_001004065
NM_001198656
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr6_+_30959839
|
2.206
|
NM_001954
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr1_-_151866593
|
2.200
|
NM_001024211
|
S100A13
|
S100 calcium binding protein A13
|
|
chr12_+_74160779
|
2.197
|
NM_006851
|
GLIPR1
|
GLI pathogenesis-related 1
|
|
chr1_-_205272619
|
2.195
|
NM_001083924
NM_023938
|
C1orf116
|
chromosome 1 open reading frame 116
|
|
chr5_-_156935317
|
2.153
|
NM_033274
|
ADAM19
|
ADAM metallopeptidase domain 19
|
|
chr10_+_89409332
|
2.114
|
NM_001015880
NM_004670
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2
|
|
chr9_+_115382760
|
2.105
|
NM_021106
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr11_-_46678691
|
2.045
|
NM_004308
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr5_+_150380325
|
2.041
|
|
GPX3
|
glutathione peroxidase 3 (plasma)
|
|
chr9_-_73573121
|
1.974
|
NM_001135820
NM_013390
|
TMEM2
|
transmembrane protein 2
|
|
chr11_-_11987115
|
1.973
|
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr8_-_119192985
|
1.965
|
NM_000127
|
EXT1
|
exostosin 1
|
|
chr3_+_139550368
|
1.951
|
|
MRAS
|
muscle RAS oncogene homolog
|
|
chr6_+_86216457
|
1.943
|
|
NT5E
|
5'-nucleotidase, ecto (CD73)
|
|
chr1_-_68470585
|
1.928
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chr5_+_150380270
|
1.904
|
|
GPX3
|
glutathione peroxidase 3 (plasma)
|
|
chr4_+_146622154
|
1.887
|
NM_005900
|
SMAD1
|
SMAD family member 1
|
|
chr5_+_150380173
|
1.886
|
NM_002084
|
GPX3
|
glutathione peroxidase 3 (plasma)
|
|
chr2_-_105381855
|
1.871
|
NM_001039492
NM_001450
NM_201555
|
FHL2
|
four and a half LIM domains 2
|
|
chr4_+_75699652
|
1.844
|
NM_001657
|
AREG
|
amphiregulin
|
|
chrX_-_68301974
|
1.820
|
NM_001032396
NM_022368
NM_145119
|
PJA1
|
praja ring finger 1
|
|
chr15_+_65145248
|
1.815
|
NM_005902
|
SMAD3
|
SMAD family member 3
|
|
chr1_-_94779727
|
1.813
|
NM_001178096
NM_001993
|
F3
|
coagulation factor III (thromboplastin, tissue factor)
|
|
chr6_-_150285838
|
1.806
|
NM_001001788
|
RAET1G
|
retinoic acid early transcript 1G
|
|
chr22_+_37193961
|
1.782
|
NM_006855
NM_016657
|
KDELR3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3
|
|
chr5_+_34720368
|
1.774
|
NM_001145521
NM_001145525
|
RAI14
|
retinoic acid induced 14
|
|
chr9_+_114953087
|
1.771
|
|
SLC31A2
|
solute carrier family 31 (copper transporters), member 2
|
|
chr2_+_37425240
|
1.730
|
NM_012413
|
QPCT
|
glutaminyl-peptide cyclotransferase
|
|
chr8_-_11762918
|
1.670
|
|
CTSB
|
cathepsin B
|
|
chr8_-_11763004
|
1.670
|
|
CTSB
|
cathepsin B
|
|
chr12_+_54611212
|
1.667
|
NM_001345
|
DGKA
|
diacylglycerol kinase, alpha 80kDa
|
|
chr12_+_54612055
|
1.659
|
NM_201444
NM_201445
NM_201554
|
DGKA
|
diacylglycerol kinase, alpha 80kDa
|
|
chr9_-_112840801
|
1.646
|
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr20_+_17498507
|
1.621
|
NM_001011546
NM_006870
|
DSTN
|
destrin (actin depolymerizing factor)
|
|
chr4_+_158216631
|
1.616
|
NM_000824
NM_001166061
NM_001166060
|
GLRB
|
glycine receptor, beta
|
|
chr15_+_65145215
|
1.613
|
|
SMAD3
|
SMAD family member 3
|
|
chr11_-_11987198
|
1.613
|
NM_015881
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr9_+_115382990
|
1.607
|
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr14_-_23734598
|
1.594
|
NM_001014842
NM_006405
|
TM9SF1
|
transmembrane 9 superfamily member 1
|
|
chr8_-_11763023
|
1.580
|
|
CTSB
|
cathepsin B
|
|
chr13_+_47705308
|
1.573
|
|
ITM2B
|
integral membrane protein 2B
|
|
chr17_+_35097851
|
1.548
|
NM_001005862
|
ERBB2
|
v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian)
|
|
chr19_-_63758115
|
1.496
|
|
CHMP2A
|
chromatin modifying protein 2A
|
|
chr1_-_151855413
|
1.495
|
NM_020672
|
S100A14
|
S100 calcium binding protein A14
|
|
chrX_+_154264931
|
1.486
|
NM_001007523
NM_001007524
NM_012151
|
F8A2
F8A3
F8A1
|
coagulation factor VIII-associated (intronic transcript) 2
coagulation factor VIII-associated (intronic transcript) 3
coagulation factor VIII-associated (intronic transcript) 1
|
|
chr11_-_46678715
|
1.475
|
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr10_-_725522
|
1.455
|
NM_014974
|
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila)
|
|
chr19_-_63758165
|
1.452
|
NM_198426
|
CHMP2A
|
chromatin modifying protein 2A
|
|
chr11_-_46678736
|
1.447
|
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr19_-_63758184
|
1.435
|
|
CHMP2A
|
chromatin modifying protein 2A
|
|
chr17_-_74433048
|
1.420
|
NM_003255
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr3_-_50371934
|
1.412
|
NM_007024
|
TMEM115
|
transmembrane protein 115
|
|
chrX_-_151749915
|
1.405
|
NM_004344
|
CETN2
|
centrin, EF-hand protein, 2
|
|
chr4_+_148621518
|
1.396
|
NM_001166055
NM_001957
|
EDNRA
|
endothelin receptor type A
|
|
chr3_-_50371896
|
1.383
|
|
TMEM115
|
transmembrane protein 115
|
|
chr19_-_63758270
|
1.379
|
|
CHMP2A
|
chromatin modifying protein 2A
|
|
chr9_+_123123364
|
1.367
|
|
GSN
|
gelsolin
|
|
chr19_-_50619015
|
1.363
|
NM_001166049
NM_001983
|
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence)
|
|
chr19_-_50618659
|
1.358
|
NM_202001
|
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence)
|
|
chr3_-_50371960
|
1.355
|
|
TMEM115
|
transmembrane protein 115
|
|
chr9_+_132310068
|
1.340
|
|
ASS1
|
argininosuccinate synthase 1
|
|
chr9_-_129656830
|
1.334
|
NM_000118
NM_001114753
|
ENG
|
endoglin
|
|
chr14_-_23734433
|
1.328
|
|
TM9SF1
|
transmembrane 9 superfamily member 1
|
|
chr19_-_50618599
|
1.317
|
|
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence)
|
|
chr9_-_129656581
|
1.317
|
|
ENG
|
endoglin
|
|
chr4_-_111777584
|
1.315
|
|
PITX2
|
paired-like homeodomain 2
|
|
chr15_-_75150177
|
1.312
|
|
TSPAN3
|
tetraspanin 3
|
|
chr14_-_23734446
|
1.310
|
|
TM9SF1
|
transmembrane 9 superfamily member 1
|
|
chr19_-_63758289
|
1.296
|
NM_014453
|
CHMP2A
|
chromatin modifying protein 2A
|
|
chr19_-_50618921
|
1.294
|
|
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence)
|
|
chr9_-_32516164
|
1.285
|
NM_014314
|
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58
|
|
chr18_-_20231783
|
1.266
|
NM_080597
|
OSBPL1A
|
oxysterol binding protein-like 1A
|
|
chr19_-_50618982
|
1.263
|
|
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence)
|
|
chr21_-_43671355
|
1.254
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr9_+_35595339
|
1.246
|
|
TESK1
|
testis-specific kinase 1
|
|
chr17_-_43470134
|
1.243
|
NM_016429
|
COPZ2
|
coatomer protein complex, subunit zeta 2
|
|
chr3_+_132228383
|
1.237
|
NM_001146003
NM_024800
NM_145910
|
NEK11
|
NIMA (never in mitosis gene a)- related kinase 11
|
|
chr5_+_141468568
|
1.224
|
|
NDFIP1
|
Nedd4 family interacting protein 1
|
|
chr4_-_111777651
|
1.216
|
|
PITX2
|
paired-like homeodomain 2
|
|
chr1_+_208569262
|
1.215
|
NM_001170580
|
HHAT
|
hedgehog acyltransferase
|
|
chr3_+_156280604
|
1.213
|
NM_007288
|
MME
|
membrane metallo-endopeptidase
|
|
chr3_+_191714533
|
1.211
|
NM_001167928
NM_001167929
NM_001167930
NM_001167931
NM_002182
NM_134470
|
IL1RAP
|
interleukin 1 receptor accessory protein
|
|
chr15_-_68971665
|
1.203
|
NM_020147
|
THAP10
|
THAP domain containing 10
|
|
chr5_+_141468498
|
1.196
|
NM_030571
|
NDFIP1
|
Nedd4 family interacting protein 1
|
|
chr11_-_61491472
|
1.177
|
|
FTH1
|
ferritin, heavy polypeptide 1
|
|
chr1_-_151866367
|
1.173
|
NM_001024212
|
S100A13
|
S100 calcium binding protein A13
|
|
chr1_+_226394605
|
1.171
|
|
GUK1
|
guanylate kinase 1
|
|
chr9_+_35595275
|
1.150
|
NM_006285
|
TESK1
|
testis-specific kinase 1
|
|
chr9_-_129656778
|
1.150
|
|
ENG
|
endoglin
|
|
chr1_-_109386112
|
1.145
|
NM_001142550
NM_001142551
NM_014969
|
WDR47
|
WD repeat domain 47
|
|
chr1_+_77105709
|
1.144
|
|
ST6GALNAC5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5
|
|
chr1_+_226394626
|
1.142
|
|
GUK1
|
guanylate kinase 1
|
|
chr4_+_6627801
|
1.138
|
NM_015274
|
MAN2B2
|
mannosidase, alpha, class 2B, member 2
|
|
chr9_-_100057809
|
1.124
|
NM_018421
|
TBC1D2
|
TBC1 domain family, member 2
|
|
chr3_-_49916304
|
1.118
|
NM_002447
|
MST1R
|
macrophage stimulating 1 receptor (c-met-related tyrosine kinase)
|
|
chr1_+_77105746
|
1.116
|
NM_030965
|
ST6GALNAC5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5
|
|
chr1_-_6243621
|
1.115
|
NM_207370
|
GPR153
|
G protein-coupled receptor 153
|
|
chr8_-_11763037
|
1.112
|
NM_001908
NM_147780
NM_147781
NM_147782
NM_147783
|
CTSB
|
cathepsin B
|
|
chr1_+_226394638
|
1.103
|
|
GUK1
|
guanylate kinase 1
|
|
chr19_+_52908412
|
1.091
|
NM_014601
|
EHD2
|
EH-domain containing 2
|
|
chr9_-_129656413
|
1.088
|
|
ENG
|
endoglin
|
|
chr5_-_122400219
|
1.080
|
|
PPIC
|
peptidylprolyl isomerase C (cyclophilin C)
|
|
chr14_+_58174489
|
1.075
|
NM_001079520
NM_016651
|
DACT1
|
dapper, antagonist of beta-catenin, homolog 1 (Xenopus laevis)
|
|
chr4_+_148621567
|
1.073
|
|
EDNRA
|
endothelin receptor type A
|
|
chr15_-_58477407
|
1.070
|
NM_001002857
NM_001002858
NM_001136015
NM_004039
|
ANXA2
|
annexin A2
|
|
chr1_+_226394545
|
1.057
|
NM_000858
NM_001159390
|
GUK1
|
guanylate kinase 1
|
|
chr4_+_38545741
|
1.055
|
NM_138389
|
FAM114A1
|
family with sequence similarity 114, member A1
|
|
chr5_+_141468569
|
1.053
|
|
NDFIP1
|
Nedd4 family interacting protein 1
|
|
chr19_+_52973653
|
1.049
|
NM_003009
|
SEPW1
|
selenoprotein W, 1
|
|
chr15_+_65217412
|
1.034
|
NM_001145103
|
SMAD3
|
SMAD family member 3
|
|
chrX_+_118254233
|
1.033
|
NM_006667
|
PGRMC1
|
progesterone receptor membrane component 1
|
|
chr5_+_96024434
|
1.025
|
|
CAST
|
calpastatin
|
|
chr17_-_78249750
|
1.024
|
NM_006822
|
RAB40B
|
RAB40B, member RAS oncogene family
|
|
chr5_+_140777454
|
1.013
|
NM_018927
NM_032101
|
PCDHGB7
|
protocadherin gamma subfamily B, 7
|
|
chr11_+_93501791
|
1.002
|
|
PANX1
|
pannexin 1
|
|
chr20_+_42777419
|
1.001
|
|
WISP2
|
WNT1 inducible signaling pathway protein 2
|
|
chr1_+_226394589
|
0.994
|
|
GUK1
|
guanylate kinase 1
|
|
chr6_-_139737477
|
0.991
|
NM_006079
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr9_+_129951552
|
0.990
|
NM_005564
|
LCN2
|
lipocalin 2
|
|
chr1_-_94475504
|
0.970
|
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr9_-_100057313
|
0.968
|
|
TBC1D2
|
TBC1 domain family, member 2
|
|
chr4_+_15313789
|
0.963
|
|
BST1
|
bone marrow stromal cell antigen 1
|
|
chr4_+_6627816
|
0.946
|
|
MAN2B2
|
mannosidase, alpha, class 2B, member 2
|
|
chr11_+_93501708
|
0.945
|
NM_015368
|
PANX1
|
pannexin 1
|
|
chr5_+_15553288
|
0.927
|
NM_012304
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chr3_+_139550134
|
0.926
|
NM_001085049
|
MRAS
|
muscle RAS oncogene homolog
|
|
chr17_+_19221606
|
0.923
|
NM_139032
NM_139033
|
MAPK7
|
mitogen-activated protein kinase 7
|
|
chr15_-_56829069
|
0.920
|
|
ADAM10
|
ADAM metallopeptidase domain 10
|
|
chr8_+_22493928
|
0.919
|
NM_176871
NM_198042
|
PDLIM2
|
PDZ and LIM domain 2 (mystique)
|
|
chr19_-_50618471
|
0.914
|
|
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence)
|
|
chr17_-_53335748
|
0.908
|
NM_017949
|
CUEDC1
|
CUE domain containing 1
|
|
chr6_-_139737432
|
0.905
|
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr8_-_54918325
|
0.903
|
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr18_+_12298189
|
0.901
|
|
TUBB6
|
tubulin, beta 6
|
|
chr16_+_65765256
|
0.898
|
NM_003946
NM_001185057
|
NOL3
|
nucleolar protein 3 (apoptosis repressor with CARD domain)
|
|
chr12_-_100979923
|
0.889
|
NM_016053
|
CCDC53
|
coiled-coil domain containing 53
|
|
chr14_+_89934358
|
0.882
|
NM_001166106
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta)
|
|
chr8_-_11763031
|
0.879
|
|
CTSB
|
cathepsin B
|
|
chr2_-_55131096
|
0.874
|
|
RTN4
|
reticulon 4
|
|
chr8_+_1759502
|
0.874
|
NM_014629
|
ARHGEF10
|
Rho guanine nucleotide exchange factor (GEF) 10
|
|
chr19_-_4351357
|
0.873
|
|
SH3GL1
|
SH3-domain GRB2-like 1
|
|
chr10_+_17311282
|
0.869
|
|
VIM
|
vimentin
|
|
chr8_+_79740813
|
0.868
|
NM_016010
|
FAM164A
|
family with sequence similarity 164, member A
|
|
chr21_-_42247007
|
0.866
|
NM_015500
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr19_+_54722686
|
0.866
|
NM_020650
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr1_+_201862552
|
0.859
|
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4
|
|
chr1_+_208568811
|
0.845
|
NM_001170587
NM_001170588
NM_018194
|
HHAT
|
hedgehog acyltransferase
|
|
chr8_-_54918369
|
0.837
|
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr11_-_61491643
|
0.833
|
|
FTH1
CPSF7
|
ferritin, heavy polypeptide 1
cleavage and polyadenylation specific factor 7, 59kDa
|
|
chr14_-_74148422
|
0.831
|
NM_000428
|
LTBP2
|
latent transforming growth factor beta binding protein 2
|