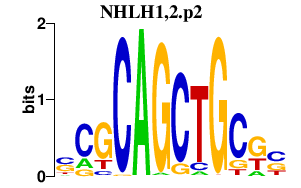
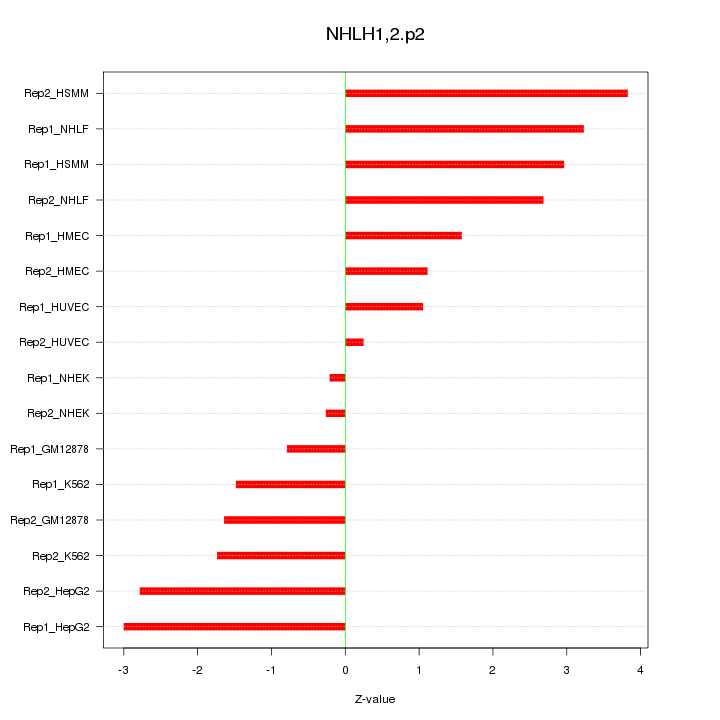
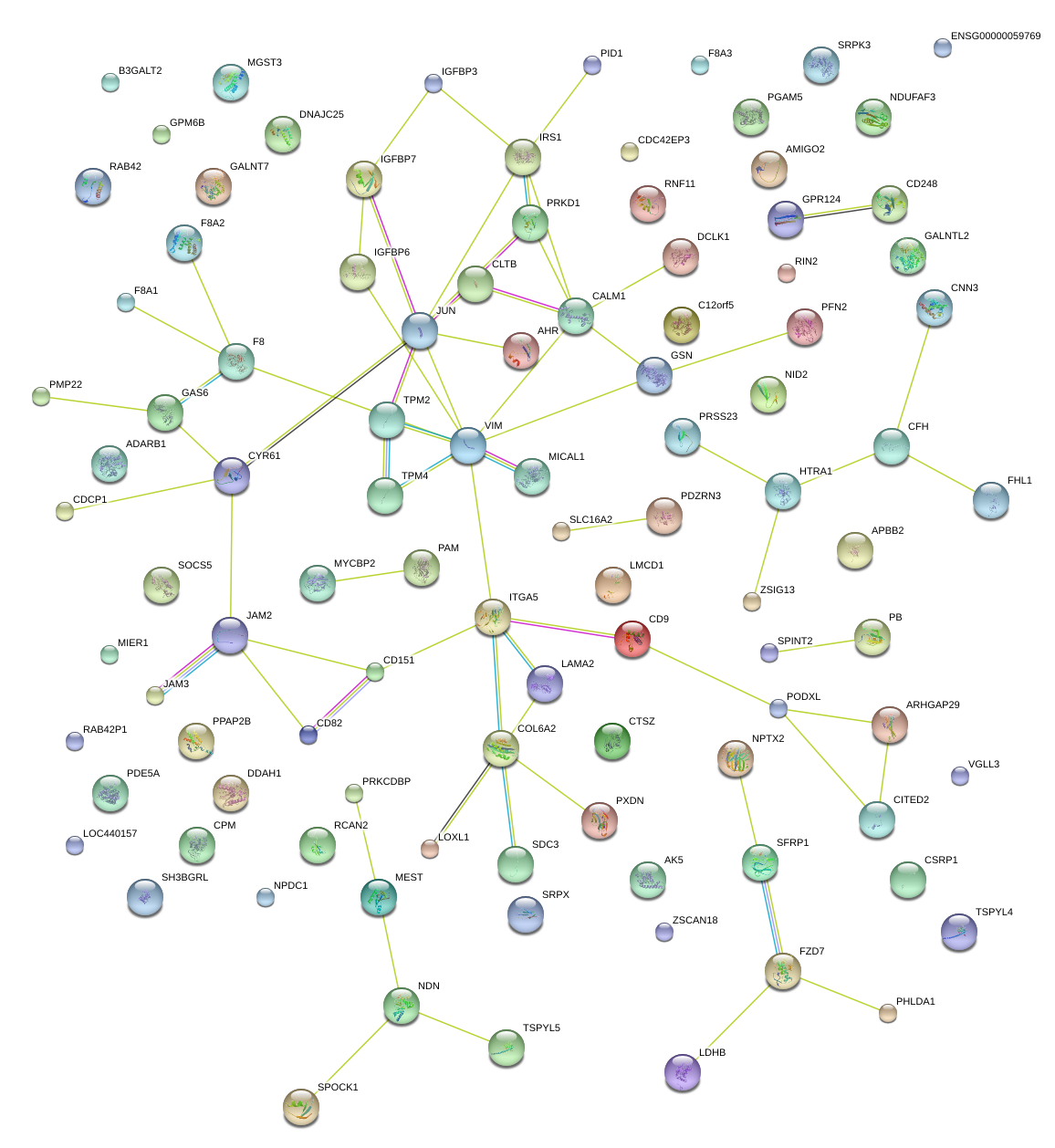
|
chr13_+_113546841
|
4.628
|
NM_000820
|
GAS6
|
growth arrest-specific 6
|
|
chr10_+_17311283
|
4.281
|
|
VIM
|
vimentin
|
|
chr10_+_17311307
|
4.264
|
|
VIM
|
vimentin
|
|
chr10_+_17311242
|
4.254
|
|
VIM
|
vimentin
|
|
chr10_+_17311282
|
4.127
|
|
VIM
|
vimentin
|
|
chr10_+_17311347
|
4.118
|
|
VIM
|
vimentin
|
|
chr13_+_113546897
|
3.953
|
|
GAS6
|
growth arrest-specific 6
|
|
chr14_-_51605487
|
3.918
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr13_+_113546943
|
3.836
|
|
GAS6
|
growth arrest-specific 6
|
|
chrX_-_13866451
|
3.786
|
|
GPM6B
|
glycoprotein M6B
|
|
chr4_-_57671307
|
3.689
|
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr11_-_6298284
|
3.581
|
NM_145040
|
PRKCDBP
|
protein kinase C, delta binding protein
|
|
chr14_-_51605461
|
3.561
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr14_-_51605515
|
3.448
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr9_-_35679799
|
3.328
|
|
TPM2
|
tropomyosin 2 (beta)
|
|
chr2_-_229844198
|
3.311
|
NM_001100818
NM_017933
|
PID1
|
phosphotyrosine interaction domain containing 1
|
|
chr4_-_57671273
|
3.149
|
NM_001553
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr9_-_35679902
|
3.088
|
NM_003289
NM_213674
|
TPM2
|
tropomyosin 2 (beta)
|
|
chr10_+_124211353
|
2.990
|
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr7_+_129919157
|
2.825
|
NM_002402
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr4_-_81213207
|
2.750
|
|
|
|
|
chr2_-_37752731
|
2.612
|
NM_006449
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr2_-_1727247
|
2.520
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr12_-_21702021
|
2.491
|
|
LDHB
|
lactate dehydrogenase B
|
|
chr10_+_17311788
|
2.478
|
|
VIM
|
vimentin
|
|
chr12_-_21702000
|
2.471
|
|
LDHB
|
lactate dehydrogenase B
|
|
chr12_-_21702036
|
2.422
|
|
LDHB
|
lactate dehydrogenase B
|
|
chr11_+_86189138
|
2.361
|
NM_007173
|
PRSS23
|
protease, serine, 23
|
|
chr11_-_65841077
|
2.349
|
NM_020404
|
CD248
|
CD248 molecule, endosialin
|
|
chr13_+_113546912
|
2.342
|
|
GAS6
|
growth arrest-specific 6
|
|
chr4_-_40911212
|
2.318
|
NM_001166050
NM_004307
NM_173075
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr15_+_72006015
|
2.293
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr3_-_73756668
|
2.290
|
NM_015009
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr12_-_53099269
|
2.262
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr12_-_53099203
|
2.157
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr3_-_151171576
|
2.089
|
|
PFN2
|
profilin 2
|
|
chr1_+_67168470
|
2.043
|
NM_001077701
NM_001077704
|
MIER1
|
mesoderm induction early response 1 homolog (Xenopus laevis)
|
|
chr7_-_130891861
|
2.040
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr7_-_45927263
|
2.000
|
NM_000598
NM_001013398
|
IGFBP3
|
insulin-like growth factor binding protein 3
|
|
chr7_+_17304707
|
1.975
|
NM_001621
|
AHR
|
aryl hydrocarbon receptor
|
|
chr12_-_53099277
|
1.940
|
NM_002205
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr19_+_16048047
|
1.925
|
NM_003290
|
TPM4
|
tropomyosin 4
|
|
chr1_+_28791593
|
1.889
|
NM_001193532
|
RAB42
|
RAB42, member RAS oncogene family
|
|
chr14_-_51605695
|
1.861
|
NM_007361
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr1_+_77520315
|
1.851
|
NM_174858
|
AK5
|
adenylate kinase 5
|
|
chr14_+_89933118
|
1.819
|
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta)
|
|
chrX_-_13866565
|
1.815
|
NM_001001994
|
GPM6B
|
glycoprotein M6B
|
|
chr1_-_85703321
|
1.813
|
NM_012137
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr12_+_6179736
|
1.738
|
NM_001769
|
CD9
|
CD9 molecule
|
|
chr12_+_51777684
|
1.631
|
NM_002178
|
IGFBP6
|
insulin-like growth factor binding protein 6
|
|
chr13_-_35603432
|
1.626
|
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr21_+_46367216
|
1.576
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr11_+_133444175
|
1.572
|
|
JAM3
|
junctional adhesion molecule 3
|
|
chr3_-_151171190
|
1.530
|
NM_002628
NM_053024
|
PFN2
|
profilin 2
|
|
chr12_-_45759691
|
1.525
|
NM_001143668
NM_181847
|
AMIGO2
|
adhesion molecule with Ig-like domain 2
|
|
chr1_-_95165089
|
1.524
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr19_-_63301418
|
1.523
|
NM_001145543
NM_001145544
NM_023926
|
ZSCAN18
|
zinc finger and SCAN domain containing 18
|
|
chr1_+_85819004
|
1.519
|
NM_001554
|
CYR61
|
cysteine-rich, angiogenic inducer, 61
|
|
chr17_-_15106597
|
1.501
|
NM_153321
|
PMP22
|
peripheral myelin protein 22
|
|
chr3_-_87122936
|
1.498
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr17_-_15106579
|
1.493
|
|
PMP22
|
peripheral myelin protein 22
|
|
chrX_-_154341451
|
1.482
|
NM_001007523
NM_001007524
NM_012151
|
F8A2
F8A3
F8A1
|
coagulation factor VIII-associated (intronic transcript) 2
coagulation factor VIII-associated (intronic transcript) 3
coagulation factor VIII-associated (intronic transcript) 1
|
|
chr10_+_124210980
|
1.477
|
NM_002775
|
HTRA1
|
HtrA serine peptidase 1
|
|
chrX_+_135057202
|
1.469
|
NM_001159702
NM_001159703
NM_001449
|
FHL1
|
four and a half LIM domains 1
|
|
chr9_-_139060416
|
1.462
|
NM_015392
|
NPDC1
|
neural proliferation, differentiation and control, 1
|
|
chr5_-_136862786
|
1.460
|
NM_004598
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr4_+_174326464
|
1.432
|
NM_017423
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7)
|
|
chrX_+_153767803
|
1.430
|
NM_001007523
NM_001007524
NM_012151
|
F8A2
F8A3
F8A1
|
coagulation factor VIII-associated (intronic transcript) 2
coagulation factor VIII-associated (intronic transcript) 3
coagulation factor VIII-associated (intronic transcript) 1
|
|
chr3_+_8518508
|
1.424
|
NM_014583
|
LMCD1
|
LIM and cysteine-rich domains 1
|
|
chr2_-_227371698
|
1.397
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr12_-_74710741
|
1.381
|
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr1_-_191422346
|
1.379
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr19_+_43446937
|
1.367
|
NM_001166103
NM_021102
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chrX_-_37964924
|
1.352
|
NM_001170750
NM_001170751
NM_001170752
NM_006307
|
SRPX
|
sushi-repeat containing protein, X-linked
|
|
chr8_-_98359248
|
1.316
|
|
TSPYL5
|
TSPY-like 5
|
|
chr1_-_31154023
|
1.307
|
NM_014654
|
SDC3
|
syndecan 3
|
|
chr2_-_1727324
|
1.301
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr8_-_41286088
|
1.298
|
NM_003012
|
SFRP1
|
secreted frizzled-related protein 1
|
|
chr20_+_19818209
|
1.296
|
NM_018993
|
RIN2
|
Ras and Rab interactor 2
|
|
chr19_+_43447078
|
1.295
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr7_+_98084531
|
1.292
|
NM_002523
|
NPTX2
|
neuronal pentraxin II
|
|
chr11_+_44543716
|
1.275
|
NM_001024844
NM_002231
|
CD82
|
CD82 molecule
|
|
chr5_+_102229612
|
1.237
|
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr14_+_89933055
|
1.233
|
NM_006888
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta)
|
|
chr2_-_1727292
|
1.229
|
NM_012293
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr1_-_59020875
|
1.227
|
|
JUN
|
jun proto-oncogene
|
|
chr3_+_49034494
|
1.214
|
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 3
|
|
chr9_+_123070234
|
1.210
|
|
GSN
|
gelsolin
|
|
chr6_-_109883635
|
1.201
|
|
MICAL1
|
microtubule associated monoxygenase, calponin and LIM domain containing 1
|
|
chr1_-_56817717
|
1.200
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr8_-_98359347
|
1.186
|
NM_033512
|
TSPYL5
|
TSPY-like 5
|
|
chrX_-_13866366
|
1.177
|
|
|
|
|
chr6_-_109883639
|
1.177
|
|
MICAL1
|
microtubule associated monoxygenase, calponin and LIM domain containing 1
|
|
chr12_+_4300607
|
1.173
|
NM_020375
|
C12orf5
|
chromosome 12 open reading frame 5
|
|
chr1_+_163867057
|
1.168
|
NM_004528
|
MGST3
|
microsomal glutathione S-transferase 3
|
|
chr3_-_45162865
|
1.165
|
|
CDCP1
|
CUB domain containing protein 1
|
|
chr20_-_57015698
|
1.165
|
NM_001336
|
CTSZ
|
cathepsin Z
|
|
chr5_-_175776137
|
1.164
|
NM_001834
NM_007097
|
CLTB
|
clathrin, light chain B
|
|
chrX_+_80344253
|
1.158
|
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chr1_+_51474531
|
1.158
|
NM_014372
|
RNF11
|
ring finger protein 11
|
|
chr6_+_129245978
|
1.158
|
NM_000426
NM_001079823
|
LAMA2
|
laminin, alpha 2
|
|
chr19_+_16048266
|
1.150
|
|
TPM4
|
tropomyosin 4
|
|
chr19_+_16048306
|
1.149
|
|
TPM4
|
tropomyosin 4
|
|
chr6_-_46566969
|
1.147
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chr3_+_49034050
|
1.141
|
NM_199069
NM_199070
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 3
|
|
chr4_-_120769275
|
1.141
|
NM_001083
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific
|
|
chr2_+_46779827
|
1.140
|
NM_144949
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr6_-_139737432
|
1.117
|
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr12_-_67613213
|
1.114
|
NM_001005502
NM_198320
|
CPM
|
carboxypeptidase M
|
|
chr1_-_94475840
|
1.114
|
NM_004815
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chrX_+_152699616
|
1.109
|
NM_001170760
NM_001170761
NM_014370
|
SRPK3
|
SRSF protein kinase 3
|
|
chr6_-_139737477
|
1.106
|
NM_006079
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr19_+_16048315
|
1.105
|
|
TPM4
|
tropomyosin 4
|
|
chr3_-_45162901
|
1.089
|
NM_022842
NM_178181
|
CDCP1
|
CUB domain containing protein 1
|
|
chr11_+_822965
|
1.085
|
|
CD151
|
CD151 molecule (Raph blood group)
|
|
chr2_+_202607554
|
1.077
|
NM_003507
|
FZD7
|
frizzled homolog 7 (Drosophila)
|
|
chr8_+_37773991
|
1.073
|
|
GPR124
|
G protein-coupled receptor 124
|
|
chr1_-_199732323
|
1.064
|
NM_001193572
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr11_+_823007
|
1.060
|
|
CD151
|
CD151 molecule (Raph blood group)
|
|
chr11_+_822909
|
1.060
|
NM_001039490
NM_004357
NM_139029
NM_139030
|
CD151
|
CD151 molecule (Raph blood group)
|
|
chr9_+_113433364
|
1.046
|
NM_001015882
NM_004125
|
DNAJC25
DNAJC25-GNG10
|
DnaJ (Hsp40) homolog, subfamily C , member 25
DNAJC25-GNG10 readthrough
|
|
chrX_+_73557809
|
1.042
|
NM_006517
|
SLC16A2
|
solute carrier family 16, member 2 (monocarboxylic acid transporter 8)
|
|
chr3_-_45162847
|
1.039
|
|
CDCP1
|
CUB domain containing protein 1
|
|
chr8_+_37773564
|
1.039
|
|
GPR124
|
G protein-coupled receptor 124
|
|
chr8_+_37773915
|
1.037
|
|
GPR124
|
G protein-coupled receptor 124
|
|
chr21_+_25933459
|
1.030
|
NM_021219
|
JAM2
|
junctional adhesion molecule 2
|
|
chr21_+_45318861
|
1.025
|
NM_001112
NM_001160230
NM_015833
NM_015834
|
ADARB1
|
adenosine deaminase, RNA-specific, B1
|
|
chr15_-_21483457
|
1.020
|
|
NDN
|
necdin homolog (mouse)
|
|
chr14_-_29466562
|
1.018
|
NM_002742
|
PRKD1
|
protein kinase D1
|
|
chr7_+_65307816
|
1.015
|
|
TPST1
|
tyrosylprotein sulfotransferase 1
|
|
chr12_-_102968044
|
1.013
|
NM_031302
|
GLT8D2
|
glycosyltransferase 8 domain containing 2
|
|
chr6_-_109883525
|
0.996
|
|
MICAL1
|
microtubule associated monoxygenase, calponin and LIM domain containing 1
|
|
chr12_-_54409107
|
0.991
|
NM_001040034
NM_001780
|
CD63
|
CD63 molecule
|
|
chr12_-_102967828
|
0.983
|
|
GLT8D2
|
glycosyltransferase 8 domain containing 2
|
|
chr9_+_89530896
|
0.983
|
|
CTSL1
|
cathepsin L1
|
|
chr12_-_75796898
|
0.978
|
NM_001321
|
CSRP2
|
cysteine and glycine-rich protein 2
|
|
chr5_+_102229425
|
0.975
|
NM_000919
NM_001177306
NM_138766
NM_138821
NM_138822
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr11_-_102174032
|
0.973
|
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase)
|
|
chr6_-_139737042
|
0.969
|
NM_001168389
NM_001168388
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr1_-_94475504
|
0.969
|
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr1_+_51474583
|
0.966
|
|
RNF11
|
ring finger protein 11
|
|
chr7_-_526005
|
0.964
|
NM_002607
NM_033023
|
PDGFA
|
platelet-derived growth factor alpha polypeptide
|
|
chr9_+_89530838
|
0.962
|
|
CTSL1
|
cathepsin L1
|
|
chr12_+_47498778
|
0.962
|
NM_000725
|
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr2_-_202024472
|
0.961
|
NM_015049
|
TRAK2
|
trafficking protein, kinesin binding 2
|
|
chr7_-_124192265
|
0.959
|
|
GPR37
|
G protein-coupled receptor 37 (endothelin receptor type B-like)
|
|
chr2_+_191818124
|
0.955
|
NM_001130158
NM_012223
NM_001161819
|
MYO1B
|
myosin IB
|
|
chr13_-_109757442
|
0.953
|
NM_001845
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr1_-_154913712
|
0.952
|
NM_006617
|
NES
|
nestin
|
|
chr14_-_74148422
|
0.949
|
NM_000428
|
LTBP2
|
latent transforming growth factor beta binding protein 2
|
|
chr6_+_139497933
|
0.948
|
NM_016217
|
HECA
|
headcase homolog (Drosophila)
|
|
chr2_-_175578156
|
0.946
|
NM_001025201
NM_001822
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr4_+_78298326
|
0.945
|
|
CCNG2
|
cyclin G2
|
|
chr18_+_9324820
|
0.943
|
|
TWSG1
|
twisted gastrulation homolog 1 (Drosophila)
|
|
chr11_-_124871258
|
0.933
|
NM_005103
NM_022549
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I)
|
|
chr15_-_46725087
|
0.929
|
NM_000138
|
FBN1
|
fibrillin 1
|
|
chr5_-_175775966
|
0.925
|
|
CLTB
|
clathrin, light chain B
|
|
chr3_-_126088530
|
0.925
|
|
ITGB5
|
integrin, beta 5
|
|
chr1_-_43691932
|
0.922
|
|
HYI
|
hydroxypyruvate isomerase (putative)
|
|
chr18_+_9324762
|
0.921
|
NM_020648
|
TWSG1
|
twisted gastrulation homolog 1 (Drosophila)
|
|
chr12_-_123023312
|
0.919
|
|
CCDC92
|
coiled-coil domain containing 92
|
|
chr15_+_61127367
|
0.918
|
NM_001018008
|
TPM1
|
tropomyosin 1 (alpha)
|
|
chr13_-_44048391
|
0.917
|
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr22_+_22652248
|
0.914
|
NM_000854
|
GSTT2
GSTT2B
|
glutathione S-transferase theta 2
glutathione S-transferase theta 2B (gene/pseudogene)
|
|
chr19_+_43447262
|
0.914
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr8_+_31616809
|
0.908
|
NM_013962
|
NRG1
|
neuregulin 1
|
|
chr1_-_199613427
|
0.907
|
NM_000364
NM_001001430
NM_001001431
NM_001001432
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chr7_+_44754978
|
0.906
|
NM_031449
|
ZMIZ2
|
zinc finger, MIZ-type containing 2
|
|
chr2_-_235070431
|
0.906
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chr2_-_55131202
|
0.904
|
NM_020532
NM_153828
NM_207520
|
RTN4
|
reticulon 4
|
|
chr9_+_89530793
|
0.897
|
NM_001912
NM_145918
|
CTSL1
|
cathepsin L1
|
|
chr7_+_55054416
|
0.896
|
|
EGFR
|
epidermal growth factor receptor
|
|
chr2_+_202949289
|
0.889
|
NM_001204
|
BMPR2
|
bone morphogenetic protein receptor, type II (serine/threonine kinase)
|
|
chr9_-_139060377
|
0.889
|
|
NPDC1
|
neural proliferation, differentiation and control, 1
|
|
chrX_-_106905545
|
0.887
|
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr9_+_124173143
|
0.882
|
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr9_-_139060285
|
0.875
|
|
NPDC1
|
neural proliferation, differentiation and control, 1
|
|
chr3_-_55490401
|
0.872
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr3_-_147361465
|
0.868
|
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr13_-_35603513
|
0.866
|
NM_004734
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr12_+_77782719
|
0.866
|
|
SYT1
|
synaptotagmin I
|
|
chr1_-_201420927
|
0.864
|
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chr2_+_5750229
|
0.860
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr2_+_238060602
|
0.860
|
NM_001042467
NM_024101
|
MLPH
|
melanophilin
|
|
chr11_-_35503720
|
0.859
|
NM_001001991
NM_015430
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr4_+_158216631
|
0.859
|
NM_000824
NM_001166061
NM_001166060
|
GLRB
|
glycine receptor, beta
|
|
chr1_-_199613406
|
0.857
|
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chr4_-_57671112
|
0.845
|
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr8_+_70567581
|
0.842
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr7_+_102340579
|
0.841
|
NM_001031692
NM_005824
|
LRRC17
|
leucine rich repeat containing 17
|
|
chr5_+_17270474
|
0.841
|
NM_006317
|
BASP1
|
brain abundant, membrane attached signal protein 1
|
|
chr13_+_48967746
|
0.838
|
NM_001040443
|
PHF11
|
PHD finger protein 11
|
|
chr21_-_45118037
|
0.835
|
NM_004339
|
PTTG1IP
|
pituitary tumor-transforming 1 interacting protein
|
|
chrX_-_153767625
|
0.835
|
NM_019863
|
F8
|
coagulation factor VIII, procoagulant component
|
|
chr2_-_55130925
|
0.834
|
|
RTN4
|
reticulon 4
|
|
chr1_+_200584421
|
0.822
|
NM_001167857
NM_001167858
NM_002481
|
PPP1R12B
|
protein phosphatase 1, regulatory (inhibitor) subunit 12B
|
|
chr11_+_12355600
|
0.815
|
NM_018222
|
PARVA
|
parvin, alpha
|
|
chr1_+_84316217
|
0.813
|
NM_002731
NM_207578
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr13_+_114018459
|
0.811
|
NM_001078645
NM_003903
|
CDC16
|
cell division cycle 16 homolog (S. cerevisiae)
|
|
chr1_-_113051545
|
0.810
|
NM_001042678
NM_175744
|
RHOC
|
ras homolog gene family, member C
|