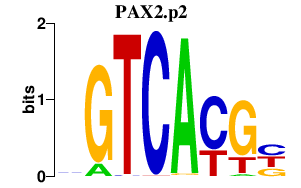
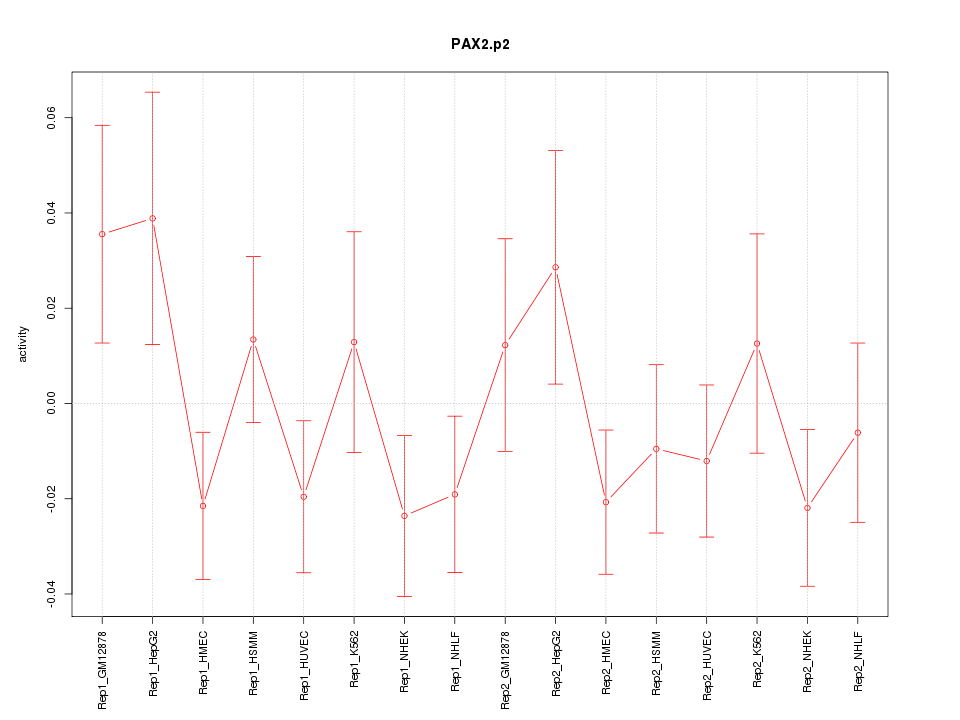
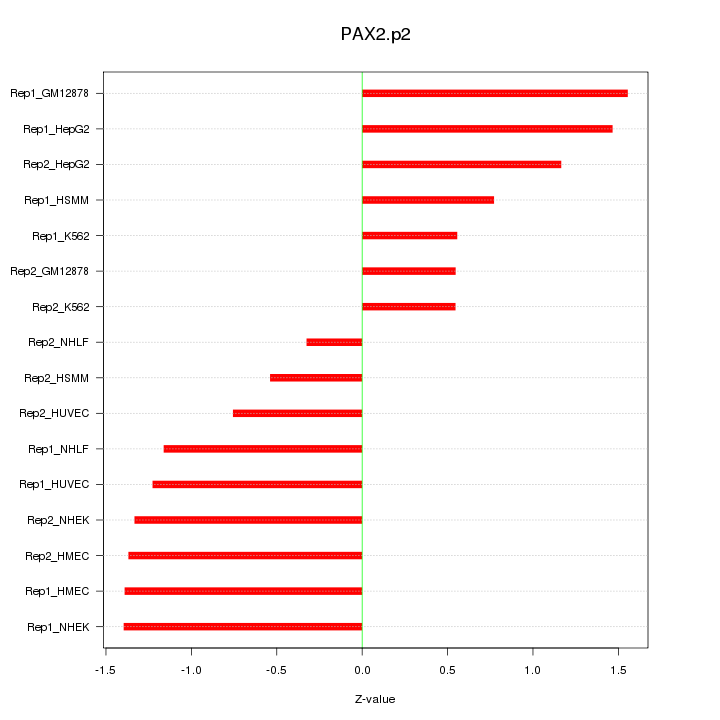
|
chr12_-_61615124
|
1.705
|
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr14_+_100270222
|
1.621
|
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chr6_+_160310118
|
1.618
|
NM_000876
|
IGF2R
|
insulin-like growth factor 2 receptor
|
|
chr12_-_61614930
|
1.480
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr17_-_39556535
|
1.298
|
NM_001015053
NM_005474
|
HDAC5
|
histone deacetylase 5
|
|
chr17_-_39556483
|
1.228
|
|
HDAC5
|
histone deacetylase 5
|
|
chr17_+_77529070
|
1.219
|
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1
|
|
chr14_+_23653848
|
1.058
|
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chrX_+_30581377
|
1.017
|
NM_000167
NM_001128127
NM_203391
|
GK
|
glycerol kinase
|
|
chr18_+_9126742
|
0.942
|
NM_001083625
NM_015208
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr14_+_23654161
|
0.924
|
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr2_+_74914742
|
0.900
|
|
HK2
|
hexokinase 2
|
|
chr17_+_75689894
|
0.897
|
NM_000152
NM_001079803
NM_001079804
|
GAA
|
glucosidase, alpha; acid
|
|
chr1_-_152459667
|
0.890
|
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chr14_+_23653819
|
0.881
|
NM_025230
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr6_-_90178685
|
0.859
|
NM_021244
|
RRAGD
|
Ras-related GTP binding D
|
|
chr11_+_7574992
|
0.835
|
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr10_+_69314347
|
0.833
|
NM_012238
|
SIRT1
|
sirtuin 1
|
|
chr14_+_23653745
|
0.831
|
NM_001163484
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr6_+_87921980
|
0.826
|
NM_015021
|
ZNF292
|
zinc finger protein 292
|
|
chr20_+_43953331
|
0.798
|
|
CTSA
|
cathepsin A
|
|
chr5_-_133734616
|
0.782
|
|
CDKL3
|
cyclin-dependent kinase-like 3
|
|
chr17_-_73636386
|
0.781
|
NM_001127198
|
TMC6
|
transmembrane channel-like 6
|
|
chr1_-_152459506
|
0.770
|
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chr16_+_23754822
|
0.766
|
|
PRKCB
|
protein kinase C, beta
|
|
chr2_+_42249896
|
0.751
|
NM_001145076
NM_019063
|
EML4
|
echinoderm microtubule associated protein like 4
|
|
chr17_+_50697355
|
0.745
|
|
HLF
|
hepatic leukemia factor
|
|
chr4_+_74498042
|
0.725
|
|
ALB
|
albumin
|
|
chr17_+_50697319
|
0.685
|
NM_002126
|
HLF
|
hepatic leukemia factor
|
|
chr20_+_43953372
|
0.685
|
|
CTSA
|
cathepsin A
|
|
chr20_+_43953428
|
0.682
|
|
CTSA
|
cathepsin A
|
|
chr6_+_138230045
|
0.681
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chrX_-_102205752
|
0.673
|
NM_018476
|
BEX1
|
brain expressed, X-linked 1
|
|
chrX_-_102205736
|
0.669
|
|
BEX1
|
brain expressed, X-linked 1
|
|
chr14_+_23654307
|
0.668
|
NM_181357
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr18_+_30875297
|
0.664
|
NM_014268
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr14_-_54438913
|
0.657
|
NM_000161
NM_001024024
NM_001024070
NM_001024071
|
GCH1
|
GTP cyclohydrolase 1
|
|
chr17_-_73636362
|
0.655
|
|
TMC6
|
transmembrane channel-like 6
|
|
chr19_-_50373294
|
0.654
|
NM_024108
|
TRAPPC6A
|
trafficking protein particle complex 6A
|
|
chr6_-_43135092
|
0.630
|
NM_015950
|
MRPL2
|
mitochondrial ribosomal protein L2
|
|
chr11_-_57039721
|
0.615
|
NM_003627
|
SLC43A1
|
solute carrier family 43, member 1
|
|
chr19_+_1018293
|
0.606
|
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr4_+_74493918
|
0.588
|
|
ALB
|
albumin
|
|
chr9_+_130749812
|
0.583
|
|
NUP188
|
nucleoporin 188kDa
|
|
chr2_+_219751192
|
0.583
|
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr19_+_1018536
|
0.575
|
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr2_+_219751230
|
0.574
|
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr17_+_77528782
|
0.572
|
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1
|
|
chr2_+_170259195
|
0.559
|
NM_144711
NM_001008489
|
PHOSPHO2-KLHL23
KLHL23
PHOSPHO2
|
PHOSPHO2-KLHL23 read-through transcript
kelch-like 23 (Drosophila)
phosphatase, orphan 2
|
|
chr16_+_23754860
|
0.559
|
|
PRKCB
|
protein kinase C, beta
|
|
chr16_+_29726962
|
0.559
|
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor)
|
|
chr2_-_44076635
|
0.558
|
NM_133259
|
LRPPRC
|
leucine-rich PPR-motif containing
|
|
chr19_+_1018114
|
0.545
|
NM_012292
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr11_+_7491789
|
0.545
|
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr11_+_120828058
|
0.543
|
NM_003105
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr5_-_175896829
|
0.543
|
NM_014901
|
RNF44
|
ring finger protein 44
|
|
chr19_-_52426055
|
0.542
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr7_+_94135384
|
0.537
|
|
PEG10
|
paternally expressed 10
|
|
chr6_-_90178630
|
0.534
|
|
RRAGD
|
Ras-related GTP binding D
|
|
chr2_-_44076616
|
0.532
|
|
LRPPRC
|
leucine-rich PPR-motif containing
|
|
chr4_+_100714995
|
0.527
|
|
MTTP
|
microsomal triglyceride transfer protein
|
|
chr17_+_75689985
|
0.526
|
|
GAA
|
glucosidase, alpha; acid
|
|
chr3_-_99724047
|
0.525
|
|
CLDND1
|
claudin domain containing 1
|
|
chr1_-_21868380
|
0.523
|
NM_001145657
NM_002885
|
RAP1GAP
|
RAP1 GTPase activating protein
|
|
chr19_-_45948211
|
0.520
|
|
C19orf54
|
chromosome 19 open reading frame 54
|
|
chr20_+_43953606
|
0.517
|
|
CTSA
|
cathepsin A
|
|
chr17_+_77528699
|
0.514
|
NM_024083
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1
|
|
chr9_-_19117494
|
0.513
|
NM_001122
|
PLIN2
|
perilipin 2
|
|
chr8_-_8788149
|
0.511
|
NM_004225
|
MFHAS1
|
malignant fibrous histiocytoma amplified sequence 1
|
|
chr11_+_65236261
|
0.511
|
|
KAT5
|
K(lysine) acetyltransferase 5
|
|
chr2_-_64734544
|
0.509
|
NM_014755
|
SERTAD2
|
SERTA domain containing 2
|
|
chr7_-_29152597
|
0.509
|
NM_019029
NM_031311
|
CPVL
|
carboxypeptidase, vitellogenic-like
|
|
chr9_+_130749791
|
0.507
|
NM_015354
|
NUP188
|
nucleoporin 188kDa
|
|
chr14_+_54807855
|
0.507
|
|
FBXO34
|
F-box protein 34
|
|
chr20_-_47965474
|
0.505
|
NM_006038
|
SPATA2
|
spermatogenesis associated 2
|
|
chr6_+_32090516
|
0.504
|
NM_007293
NM_001002029
|
C4A
C4B
|
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr1_-_11788666
|
0.500
|
NM_005957
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr6_-_90178544
|
0.499
|
|
RRAGD
|
Ras-related GTP binding D
|
|
chr11_+_61279436
|
0.498
|
NM_013279
|
C11orf9
|
chromosome 11 open reading frame 9
|
|
chr14_-_35348086
|
0.489
|
NM_014990
NM_194301
|
RALGAPA1
|
Ral GTPase activating protein, alpha subunit 1 (catalytic)
|
|
chrX_+_30581523
|
0.489
|
|
GK
|
glycerol kinase
|
|
chr14_-_34943574
|
0.488
|
NM_020529
|
NFKBIA
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha
|
|
chr6_-_29703725
|
0.487
|
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1
|
|
chr12_+_12761085
|
0.486
|
NM_004064
|
CDKN1B
|
cyclin-dependent kinase inhibitor 1B (p27, Kip1)
|
|
chr12_+_68028397
|
0.476
|
NM_000239
|
LYZ
|
lysozyme
|
|
chr4_-_155753272
|
0.471
|
|
FGG
|
fibrinogen gamma chain
|
|
chrX_-_39921525
|
0.469
|
NM_001123383
NM_001123384
|
BCOR
|
BCL6 corepressor
|
|
chr15_-_63871598
|
0.467
|
NM_001144823
NM_005848
|
DENND4A
|
DENN/MADD domain containing 4A
|
|
chr17_-_45196516
|
0.460
|
NM_030802
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr15_+_50098700
|
0.460
|
NM_002748
|
MAPK6
|
mitogen-activated protein kinase 6
|
|
chr16_+_23754772
|
0.459
|
NM_002738
NM_212535
|
PRKCB
|
protein kinase C, beta
|
|
chr17_-_23928048
|
0.456
|
NM_005165
|
ALDOC
|
aldolase C, fructose-bisphosphate
|
|
chr3_-_9895471
|
0.456
|
|
CIDEC
|
cell death-inducing DFFA-like effector c
|
|
chr6_+_64403631
|
0.455
|
|
PHF3
|
PHD finger protein 3
|
|
chr7_+_149696901
|
0.448
|
|
REPIN1
|
replication initiator 1
|
|
chr18_-_44729700
|
0.447
|
NM_001190822
|
SMAD7
|
SMAD family member 7
|
|
chr17_-_23928041
|
0.445
|
|
ALDOC
|
aldolase C, fructose-bisphosphate
|
|
chr2_+_177965617
|
0.445
|
NM_003659
|
AGPS
|
alkylglycerone phosphate synthase
|
|
chr3_-_151963607
|
0.444
|
|
SIAH2
|
seven in absentia homolog 2 (Drosophila)
|
|
chr20_+_46971618
|
0.443
|
NM_006420
|
ARFGEF2
|
ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited)
|
|
chr9_+_6747919
|
0.443
|
|
KDM4C
|
lysine (K)-specific demethylase 4C
|
|
chr3_-_151963577
|
0.437
|
|
SIAH2
|
seven in absentia homolog 2 (Drosophila)
|
|
chr10_+_69314714
|
0.435
|
|
SIRT1
|
sirtuin 1
|
|
chr1_-_27834210
|
0.435
|
NM_005248
|
FGR
|
Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog
|
|
chr18_+_30810889
|
0.434
|
NM_001143826
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr1_+_11788827
|
0.434
|
|
CLCN6
|
chloride channel 6
|
|
chr6_+_64403685
|
0.433
|
|
PHF3
|
PHD finger protein 3
|
|
chr2_+_74635354
|
0.431
|
|
DOK1
|
docking protein 1, 62kDa (downstream of tyrosine kinase 1)
|
|
chr2_-_44076543
|
0.430
|
|
LRPPRC
|
leucine-rich PPR-motif containing
|
|
chr12_-_120715937
|
0.427
|
NM_019034
|
RHOF
|
ras homolog gene family, member F (in filopodia)
|
|
chr7_+_149699185
|
0.423
|
NM_014374
|
REPIN1
|
replication initiator 1
|
|
chr11_+_7491576
|
0.423
|
NM_003621
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr15_+_57696016
|
0.418
|
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type
|
|
chr2_-_219750975
|
0.415
|
|
C2orf24
|
chromosome 2 open reading frame 24
|
|
chr1_+_40399642
|
0.414
|
|
RLF
|
rearranged L-myc fusion
|
|
chr15_-_63871492
|
0.414
|
|
DENND4A
|
DENN/MADD domain containing 4A
|
|
chr17_-_23927973
|
0.413
|
|
ALDOC
|
aldolase C, fructose-bisphosphate
|
|
chr9_-_19117462
|
0.411
|
|
PLIN2
|
perilipin 2
|
|
chr2_+_177965734
|
0.409
|
|
AGPS
|
alkylglycerone phosphate synthase
|
|
chr11_+_66915997
|
0.409
|
NM_004584
|
RAD9A
|
RAD9 homolog A (S. pombe)
|
|
chr1_-_149698516
|
0.409
|
NM_001194937
NM_001194938
NM_015100
NM_145796
|
POGZ
|
pogo transposable element with ZNF domain
|
|
chr5_+_172415891
|
0.408
|
NM_001168393
NM_001168394
NM_153607
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr22_-_35232979
|
0.408
|
|
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2
|
|
chr2_+_105320247
|
0.408
|
|
C2orf49
|
chromosome 2 open reading frame 49
|
|
chr6_-_119712596
|
0.407
|
NM_005907
|
MAN1A1
|
mannosidase, alpha, class 1A, member 1
|
|
chr10_-_64246129
|
0.405
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr6_+_64403668
|
0.403
|
|
PHF3
|
PHD finger protein 3
|
|
chr11_+_60858611
|
0.403
|
|
DAK
|
dihydroxyacetone kinase 2 homolog (S. cerevisiae)
|
|
chr1_+_40399618
|
0.402
|
NM_012421
|
RLF
|
rearranged L-myc fusion
|
|
chr20_-_47965393
|
0.402
|
|
SPATA2
|
spermatogenesis associated 2
|
|
chr4_-_155753249
|
0.398
|
|
FGG
|
fibrinogen gamma chain
|
|
chr6_+_89847183
|
0.395
|
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr12_+_68028407
|
0.390
|
|
LYZ
|
lysozyme
|
|
chr19_+_10626106
|
0.390
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr2_-_208342296
|
0.386
|
NM_003468
|
FZD5
|
frizzled homolog 5 (Drosophila)
|
|
chr21_-_42689192
|
0.383
|
NM_024022
NM_032405
|
TMPRSS3
|
transmembrane protease, serine 3
|
|
chr11_+_65236064
|
0.383
|
NM_006388
NM_182709
NM_182710
|
KAT5
|
K(lysine) acetyltransferase 5
|
|
chr6_+_89847019
|
0.382
|
NM_006813
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr10_+_76256305
|
0.382
|
NM_012330
|
MYST4
|
MYST histone acetyltransferase (monocytic leukemia) 4
|
|
chr11_-_118757574
|
0.381
|
NM_004205
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr17_+_69939130
|
0.379
|
NM_022036
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C
|
|
chr14_-_64040072
|
0.378
|
NM_006977
|
ZBTB25
|
zinc finger and BTB domain containing 25
|
|
chr4_+_74497998
|
0.377
|
|
ALB
|
albumin
|
|
chr20_+_33667160
|
0.377
|
NM_003116
|
SPAG4
|
sperm associated antigen 4
|
|
chr1_-_113300351
|
0.376
|
NM_003051
|
SLC16A1
|
solute carrier family 16, member 1 (monocarboxylic acid transporter 1)
|
|
chr16_-_23429304
|
0.375
|
NM_015044
|
GGA2
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 2
|
|
chr6_+_30632464
|
0.375
|
NM_001077497
NM_025263
|
PRR3
|
proline rich 3
|
|
chr4_-_155753287
|
0.374
|
|
FGG
|
fibrinogen gamma chain
|
|
chr5_+_133479196
|
0.372
|
NM_001134851
NM_201632
NM_201634
NM_213648
|
TCF7
|
transcription factor 7 (T-cell specific, HMG-box)
|
|
chr17_-_7078446
|
0.370
|
NM_004422
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr16_+_29726931
|
0.368
|
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor)
|
|
chr12_+_28234657
|
0.366
|
|
|
|
|
chr22_-_35233032
|
0.365
|
NM_024955
NM_001102371
|
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2
|
|
chr7_-_97339616
|
0.364
|
|
ASNS
|
asparagine synthetase (glutamine-hydrolyzing)
|
|
chr16_+_29725665
|
0.363
|
|
|
|
|
chr2_+_228045053
|
0.361
|
|
AGFG1
|
ArfGAP with FG repeats 1
|
|
chr18_+_53862888
|
0.357
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr2_+_219751145
|
0.355
|
NM_024293
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr1_-_11788550
|
0.354
|
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr4_+_38341982
|
0.354
|
NM_016531
|
KLF3
|
Kruppel-like factor 3 (basic)
|
|
chr11_-_118757477
|
0.351
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr6_-_31938530
|
0.351
|
|
NEU1
|
sialidase 1 (lysosomal sialidase)
|
|
chr5_+_94916533
|
0.351
|
NM_198150
|
ARSK
|
arylsulfatase family, member K
|
|
chr18_+_58533597
|
0.350
|
NM_194449
|
PHLPP1
|
PH domain and leucine rich repeat protein phosphatase 1
|
|
chr2_-_240613357
|
0.348
|
|
NDUFA10
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kDa
|
|
chr17_+_50699965
|
0.347
|
|
HLF
|
hepatic leukemia factor
|
|
chr16_-_5023914
|
0.347
|
|
NAGPA
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase
|
|
chr16_-_5023934
|
0.347
|
NM_016256
|
NAGPA
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase
|
|
chr2_+_86800898
|
0.346
|
NM_022780
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae)
|
|
chr17_+_52518098
|
0.345
|
|
AKAP1
|
A kinase (PRKA) anchor protein 1
|
|
chr2_-_196744546
|
0.345
|
NM_004226
|
STK17B
|
serine/threonine kinase 17b
|
|
chr5_+_137829065
|
0.345
|
|
EGR1
|
early growth response 1
|
|
chr10_-_64246080
|
0.344
|
|
EGR2
|
early growth response 2
|
|
chr6_-_109810354
|
0.341
|
|
CD164
|
CD164 molecule, sialomucin
|
|
chr5_-_167938991
|
0.341
|
NM_024594
|
PANK3
|
pantothenate kinase 3
|
|
chr3_+_187813405
|
0.340
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr14_+_67156405
|
0.338
|
|
ARG2
|
arginase, type II
|
|
chr8_+_11599119
|
0.338
|
NM_002052
|
GATA4
|
GATA binding protein 4
|
|
chr6_-_90178375
|
0.337
|
|
RRAGD
|
Ras-related GTP binding D
|
|
chr1_+_1994943
|
0.337
|
NM_001033581
|
PRKCZ
|
protein kinase C, zeta
|
|
chrX_+_128741578
|
0.333
|
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr3_-_151963653
|
0.332
|
NM_005067
|
SIAH2
|
seven in absentia homolog 2 (Drosophila)
|
|
chr4_-_23500706
|
0.332
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr10_+_104145421
|
0.330
|
NM_002502
|
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100)
|
|
chr10_+_104144218
|
0.330
|
NM_001077493
NM_001077494
|
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100)
|
|
chr14_+_22860511
|
0.329
|
|
PABPN1
|
poly(A) binding protein, nuclear 1
|
|
chr13_-_31900218
|
0.328
|
NM_001079691
NM_052818
|
N4BP2L1
|
NEDD4 binding protein 2-like 1
|
|
chr17_-_45196465
|
0.328
|
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr11_+_73036241
|
0.327
|
NM_001130034
NM_001130035
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr1_+_11788793
|
0.325
|
NM_001286
NM_021735
NM_021736
NM_021737
|
CLCN6
|
chloride channel 6
|
|
chr12_+_115660468
|
0.325
|
NM_001109903
NM_032814
|
RNFT2
|
ring finger protein, transmembrane 2
|
|
chr15_+_38843535
|
0.325
|
NM_005258
|
GCHFR
|
GTP cyclohydrolase I feedback regulator
|
|
chr14_-_90953883
|
0.325
|
NM_001080414
|
CCDC88C
|
coiled-coil domain containing 88C
|
|
chr4_-_155753207
|
0.325
|
|
FGG
|
fibrinogen gamma chain
|
|
chr15_-_68781662
|
0.324
|
NM_001008224
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr2_-_98646367
|
0.323
|
NM_001160154
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr19_+_54707787
|
0.322
|
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr16_+_23476334
|
0.321
|
NM_019116
|
UBFD1
|
ubiquitin family domain containing 1
|
|
chr7_+_149201962
|
0.321
|
|
ATP6V0E2
|
ATPase, H+ transporting V0 subunit e2
|
|
chr6_+_64403936
|
0.320
|
|
PHF3
|
PHD finger protein 3
|