|
chrX_+_43399101
|
2.947
|
|
MAOA
|
monoamine oxidase A
|
|
chr2_-_98713862
|
2.026
|
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr8_+_136538739
|
1.568
|
NM_006558
|
KHDRBS3
|
KH domain containing, RNA binding, signal transduction associated 3
|
|
chr3_-_151171190
|
1.561
|
NM_002628
NM_053024
|
PFN2
|
profilin 2
|
|
chr20_-_23566325
|
1.471
|
NM_000099
|
CST3
|
cystatin C
|
|
chr2_+_235525355
|
1.464
|
NM_014521
|
SH3BP4
|
SH3-domain binding protein 4
|
|
chr13_+_26029879
|
1.401
|
|
WASF3
|
WAS protein family, member 3
|
|
chr21_+_17807167
|
1.339
|
NM_001338
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr2_-_20288647
|
1.279
|
NM_001006946
|
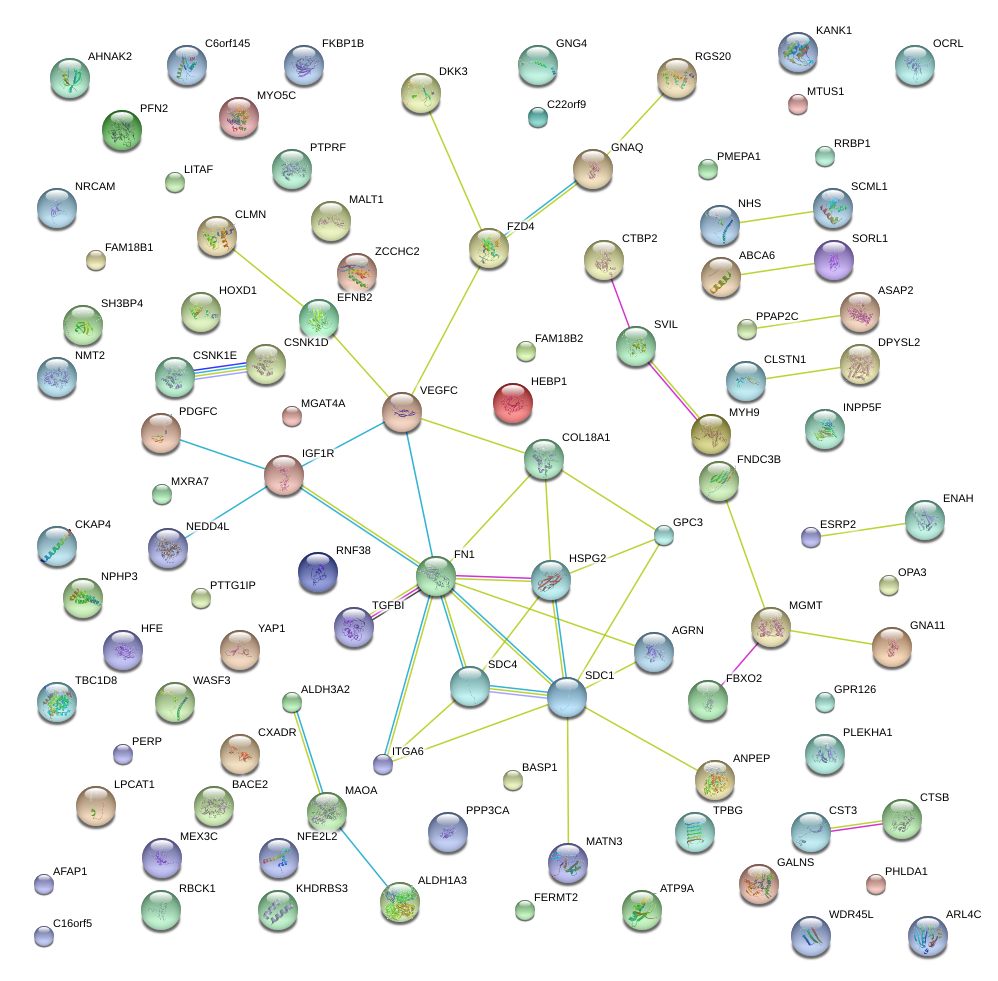
SDC1
|
syndecan 1
|
|
chr8_+_26427378
|
1.197
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr3_-_151171576
|
1.184
|
|
PFN2
|
profilin 2
|
|
chr2_-_98714020
|
1.165
|
NM_012214
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr11_+_120828058
|
1.152
|
NM_003105
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr1_-_233879617
|
1.145
|
NM_001098722
|
GNG4
|
guanine nucleotide binding protein (G protein), gamma 4
|
|
chr12_-_105165805
|
1.121
|
NM_006825
|
CKAP4
|
cytoskeleton-associated protein 4
|
|
chr14_-_104515738
|
1.104
|
NM_138420
|
AHNAK2
|
AHNAK nucleoprotein 2
|
|
chr2_-_20288108
|
1.079
|
NM_002997
|
SDC1
|
syndecan 1
|
|
chr15_+_99237527
|
1.047
|
NM_000693
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3
|
|
chr12_-_13044467
|
1.011
|
NM_015987
|
HEBP1
|
heme binding protein 1
|
|
chr21_-_45118037
|
0.999
|
NM_004339
|
PTTG1IP
|
pituitary tumor-transforming 1 interacting protein
|
|
chr15_-_50375143
|
0.998
|
NM_018728
|
MYO5C
|
myosin VC
|
|
chr10_+_124124083
|
0.977
|
NM_001001974
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr12_-_13044314
|
0.969
|
|
HEBP1
|
heme binding protein 1
|
|
chr5_+_17270474
|
0.961
|
NM_006317
|
BASP1
|
brain abundant, membrane attached signal protein 1
|
|
chr20_+_336941
|
0.957
|
|
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1
|
|
chrX_-_132947141
|
0.945
|
|
GPC3
|
glypican 3
|
|
chr22_-_44015424
|
0.934
|
|
KIAA0930
|
KIAA0930
|
|
chr13_+_26029799
|
0.925
|
NM_006646
|
WASF3
|
WAS protein family, member 3
|
|
chr9_-_36391149
|
0.925
|
NM_194330
|
RNF38
|
ring finger protein 38
|
|
chr10_+_124124196
|
0.918
|
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr5_+_17270781
|
0.918
|
|
BASP1
|
brain abundant, membrane attached signal protein 1
|
|
chrX_-_132947290
|
0.898
|
NM_001164617
NM_001164618
NM_001164619
NM_004484
|
GPC3
|
glypican 3
|
|
chr10_-_30064600
|
0.885
|
NM_003174
|
SVIL
|
supervillin
|
|
chr9_+_494661
|
0.869
|
NM_015158
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr10_+_131155443
|
0.859
|
NM_002412
|
MGMT
|
O-6-methylguanine-DNA methyltransferase
|
|
chr10_+_131155490
|
0.854
|
|
MGMT
|
O-6-methylguanine-DNA methyltransferase
|
|
chr15_+_97009194
|
0.843
|
|
IGF1R
|
insulin-like growth factor 1 receptor
|
|
chr20_-_49818306
|
0.816
|
NM_006045
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr1_+_43769065
|
0.812
|
NM_002840
NM_130440
|
PTPRF
|
protein tyrosine phosphatase, receptor type, F
|
|
chr6_-_138470160
|
0.810
|
NM_022121
|
PERP
|
PERP, TP53 apoptosis effector
|
|
chr14_-_94855848
|
0.804
|
NM_024734
|
CLMN
|
calmin (calponin-like, transmembrane)
|
|
chr20_-_49818268
|
0.803
|
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr21_-_45117989
|
0.799
|
|
PTTG1IP
|
pituitary tumor-transforming 1 interacting protein
|
|
chr5_+_135392482
|
0.784
|
NM_000358
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa
|
|
chr13_-_105985313
|
0.770
|
NM_004093
|
EFNB2
|
ephrin-B2
|
|
chr1_-_223907283
|
0.757
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr21_-_45118026
|
0.756
|
|
PTTG1IP
|
pituitary tumor-transforming 1 interacting protein
|
|
chr16_-_66827449
|
0.755
|
NM_024939
|
ESRP2
|
epithelial splicing regulatory protein 2
|
|
chr20_+_336963
|
0.754
|
|
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1
|
|
chr16_-_11588525
|
0.746
|
|
LITAF
|
lipopolysaccharide-induced TNF factor
|
|
chr10_-_15250616
|
0.739
|
NM_004808
|
NMT2
|
N-myristoyltransferase 2
|
|
chr18_+_53862888
|
0.714
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr12_-_74711642
|
0.713
|
NM_007350
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr14_-_52487386
|
0.710
|
|
FERMT2
|
fermitin family member 2
|
|
chr16_-_87450734
|
0.709
|
|
GALNS
|
galactosamine (N-acetyl)-6-sulfate sulfatase
|
|
chr1_-_11637325
|
0.703
|
NM_012168
|
FBXO2
|
F-box protein 2
|
|
chr2_-_20075889
|
0.700
|
|
MATN3
|
matrilin 3
|
|
chrX_+_17665483
|
0.698
|
NM_001037535
NM_001037536
NM_001037540
NM_006746
|
SCML1
|
sex comb on midleg-like 1 (Drosophila)
|
|
chr7_-_107883929
|
0.688
|
NM_001193582
NM_001193583
NM_001193584
NM_005010
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr21_-_45117960
|
0.686
|
|
PTTG1IP
|
pituitary tumor-transforming 1 interacting protein
|
|
chr17_-_78199384
|
0.684
|
NM_019613
|
WDR45L
|
WDR45-like
|
|
chr2_-_101134146
|
0.683
|
NM_001102426
|
TBC1D8
|
TBC1 domain family, member 8 (with GRAM domain)
|
|
chr21_+_45649429
|
0.680
|
NM_130445
|
COL18A1
|
collagen, type XVIII, alpha 1
|
|
chr6_+_29963328
|
0.675
|
|
HLA-H
|
major histocompatibility complex, class I, H (pseudogene)
|
|
chr9_-_36390817
|
0.671
|
NM_194328
NM_194332
|
RNF38
|
ring finger protein 38
|
|
chr19_-_242335
|
0.664
|
NM_003712
NM_177526
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chr10_-_126839000
|
0.658
|
|
CTBP2
|
C-terminal binding protein 2
|
|
chr12_-_74711474
|
0.657
|
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr18_+_53862744
|
0.657
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr3_+_173240983
|
0.656
|
NM_001135095
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr1_-_11637058
|
0.652
|
|
FBXO2
|
F-box protein 2
|
|
chr22_-_44015248
|
0.649
|
NM_001009880
|
KIAA0930
|
KIAA0930
|
|
chr11_-_86343859
|
0.647
|
NM_012193
|
FZD4
|
frizzled homolog 4 (Drosophila)
|
|
chr16_-_11588257
|
0.644
|
NM_001136472
NM_001136473
|
LITAF
|
lipopolysaccharide-induced TNF factor
|
|
chr20_-_43410437
|
0.642
|
NM_002999
|
SDC4
|
syndecan 4
|
|
chr10_-_126839056
|
0.639
|
NM_001329
|
CTBP2
|
C-terminal binding protein 2
|
|
chr14_-_52487558
|
0.634
|
NM_001134999
NM_001135000
NM_006832
|
FERMT2
|
fermitin family member 2
|
|
chr6_+_83130673
|
0.632
|
NM_001166392
|
TPBG
|
trophoblast glycoprotein
|
|
chr8_-_17702665
|
0.626
|
NM_001001924
NM_001001925
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr17_-_15407497
|
0.619
|
NM_001135036
NM_145301
|
FAM18B2-CDRT4
FAM18B2
|
FAM18B2-CDRT4 read-through transcript
family with sequence similarity 18, member B2
|
|
chr4_-_102487649
|
0.618
|
NM_000944
NM_001130691
NM_001130692
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme
|
|
chr1_-_9807136
|
0.617
|
NM_001009566
NM_014944
|
CLSTN1
|
calsyntenin 1
|
|
chr1_-_9806611
|
0.615
|
|
CLSTN1
|
calsyntenin 1
|
|
chr20_+_336717
|
0.614
|
|
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1
|
|
chr21_+_41461962
|
0.604
|
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chr20_-_17610704
|
0.595
|
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr3_-_133923906
|
0.593
|
NM_153240
|
NPHP3
|
nephronophthisis 3 (adolescent)
|
|
chr11_-_11987198
|
0.592
|
NM_015881
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr18_-_46978029
|
0.584
|
NM_016626
|
MEX3C
|
mex-3 homolog C (C. elegans)
|
|
chr11_-_11987115
|
0.578
|
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr15_-_88159061
|
0.577
|
NM_001150
|
ANPEP
|
alanyl (membrane) aminopeptidase
|
|
chr4_-_158111995
|
0.575
|
NM_016205
|
PDGFC
|
platelet derived growth factor C
|
|
chr18_+_54489798
|
0.570
|
|
MALT1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1
|
|
chr6_-_3697244
|
0.568
|
NM_183373
|
C6orf145
|
chromosome 6 open reading frame 145
|
|
chr17_-_72218535
|
0.561
|
NM_001008528
NM_001008529
NM_198530
|
MXRA7
|
matrix-remodelling associated 7
|
|
chr20_-_17610828
|
0.561
|
NM_001042576
NM_004587
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr1_-_22136263
|
0.561
|
NM_005529
|
HSPG2
|
heparan sulfate proteoglycan 2
|
|
chr20_-_55718351
|
0.560
|
NM_020182
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr22_-_35113923
|
0.554
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr10_+_121475628
|
0.552
|
|
INPP5F
|
inositol polyphosphate-5-phosphatase F
|
|
chr4_-_7991992
|
0.547
|
NM_001134647
NM_198595
|
AFAP1
|
actin filament associated protein 1
|
|
chr21_+_41461597
|
0.543
|
NM_012105
NM_138991
NM_138992
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chr16_-_87450849
|
0.540
|
NM_000512
|
GALNS
|
galactosamine (N-acetyl)-6-sulfate sulfatase
|
|
chr22_-_37062114
|
0.532
|
|
CSNK1E
|
casein kinase 1, epsilon
|
|
chr8_-_11763031
|
0.531
|
|
CTSB
|
cathepsin B
|
|
chr8_-_11763023
|
0.531
|
|
CTSB
|
cathepsin B
|
|
chr15_+_99237555
|
0.528
|
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3
|
|
chr1_-_233880676
|
0.525
|
NM_001098721
NM_004485
|
GNG4
|
guanine nucleotide binding protein (G protein), gamma 4
|
|
chr11_+_101485946
|
0.525
|
NM_001130145
NM_001195044
NM_006106
|
YAP1
|
Yes-associated protein 1
|
|
chr2_+_173000590
|
0.518
|
|
ITGA6
|
integrin, alpha 6
|
|
chr2_+_176761552
|
0.518
|
NM_024501
|
HOXD1
|
homeobox D1
|
|
chr2_+_9264360
|
0.516
|
|
ASAP2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2
|
|
chr22_-_35114001
|
0.511
|
NM_002473
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr8_-_11762918
|
0.509
|
|
CTSB
|
cathepsin B
|
|
chr2_+_9264327
|
0.506
|
NM_001135191
NM_003887
|
ASAP2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2
|
|
chr17_+_19492427
|
0.504
|
NM_000382
NM_001031806
|
ALDH3A2
|
aldehyde dehydrogenase 3 family, member A2
|
|
chr5_-_1577022
|
0.503
|
NM_024830
|
LPCAT1
|
lysophosphatidylcholine acyltransferase 1
|
|
chr21_+_45649719
|
0.500
|
|
|
|
|
chr2_-_216009015
|
0.496
|
NM_002026
NM_054034
NM_212474
NM_212476
NM_212478
NM_212482
|
FN1
|
fibronectin 1
|
|
chr19_+_3045528
|
0.495
|
|
GNA11
|
guanine nucleotide binding protein (G protein), alpha 11 (Gq class)
|
|
chr2_+_24126140
|
0.493
|
|
FKBP1B
|
FK506 binding protein 1B, 12.6 kDa
|
|
chr4_-_177950658
|
0.491
|
NM_005429
|
VEGFC
|
vascular endothelial growth factor C
|
|
chr18_+_54489763
|
0.491
|
|
MALT1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1
|
|
chr2_-_235070431
|
0.490
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chr8_-_11763004
|
0.490
|
|
CTSB
|
cathepsin B
|
|
chr1_+_945331
|
0.490
|
NM_198576
|
AGRN
|
agrin
|
|
chr18_+_58341637
|
0.487
|
NM_017742
|
ZCCHC2
|
zinc finger, CCHC domain containing 2
|
|
chr18_+_53862386
|
0.487
|
NM_001144967
NM_015277
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr6_+_142664490
|
0.486
|
NM_001032394
NM_001032395
NM_020455
NM_198569
|
GPR126
|
G protein-coupled receptor 126
|
|
chr16_-_4528380
|
0.475
|
NM_001199054
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr10_+_120853616
|
0.467
|
|
FAM45A
FAM45B
|
family with sequence similarity 45, member A
family with sequence similarity 45, member A pseudogene
|
|
chr4_+_6322506
|
0.466
|
|
WFS1
|
Wolfram syndrome 1 (wolframin)
|
|
chr3_+_57969127
|
0.464
|
NM_001164317
NM_001164318
NM_001164319
NM_001457
|
FLNB
|
filamin B, beta
|
|
chr1_-_11636983
|
0.464
|
|
FBXO2
|
F-box protein 2
|
|
chr5_+_52320660
|
0.464
|
NM_002203
|
ITGA2
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor)
|
|
chr6_+_138230045
|
0.461
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr6_+_29963485
|
0.459
|
|
|
|
|
chr2_-_20075929
|
0.455
|
NM_002381
|
MATN3
|
matrilin 3
|
|
chr9_+_111850698
|
0.454
|
NM_001004065
NM_001198656
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr2_-_20714303
|
0.451
|
NM_022460
|
HS1BP3
|
HCLS1 binding protein 3
|
|
chr1_-_93919789
|
0.450
|
NM_003567
|
BCAR3
|
breast cancer anti-estrogen resistance 3
|
|
chr11_+_129445057
|
0.450
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr11_+_129444997
|
0.450
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr7_+_45894480
|
0.440
|
NM_000596
|
IGFBP1
|
insulin-like growth factor binding protein 1
|
|
chr10_+_120853600
|
0.439
|
NM_207009
|
FAM45A
|
family with sequence similarity 45, member A
|
|
chr9_-_76892860
|
0.438
|
NM_001127603
NM_017881
|
C9orf95
|
chromosome 9 open reading frame 95
|
|
chr11_+_32068990
|
0.437
|
NM_002901
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain
|
|
chr17_+_45858606
|
0.434
|
|
ACSF2
|
acyl-CoA synthetase family member 2
|
|
chr1_+_9275518
|
0.434
|
NM_025106
|
SPSB1
|
splA/ryanodine receptor domain and SOCS box containing 1
|
|
chr10_+_121475583
|
0.434
|
NM_014937
|
INPP5F
|
inositol polyphosphate-5-phosphatase F
|
|
chr17_+_77779570
|
0.431
|
NM_001042422
|
SLC16A3
|
solute carrier family 16, member 3 (monocarboxylic acid transporter 4)
|
|
chr11_+_129444877
|
0.430
|
NM_001142276
NM_001142277
NM_001142278
NM_001642
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr9_+_130354490
|
0.430
|
NM_001130438
NM_001195532
NM_003127
|
SPTAN1
|
spectrin, alpha, non-erythrocytic 1 (alpha-fodrin)
|
|
chr1_-_94475504
|
0.428
|
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr2_-_224612177
|
0.423
|
NM_001136528
NM_006216
|
SERPINE2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2
|
|
chr2_+_238265519
|
0.422
|
NM_001137551
NM_001137552
NM_001137553
NM_004735
|
LRRFIP1
|
leucine rich repeat (in FLII) interacting protein 1
|
|
chrX_-_152804731
|
0.422
|
|
L1CAM
|
L1 cell adhesion molecule
|
|
chr1_+_181259317
|
0.421
|
|
|
|
|
chr15_+_67239853
|
0.418
|
NM_015554
|
GLCE
|
glucuronic acid epimerase
|
|
chr6_-_10527772
|
0.417
|
NM_001042425
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr1_-_94475840
|
0.417
|
NM_004815
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr19_-_18913032
|
0.415
|
NM_004838
|
HOMER3
|
homer homolog 3 (Drosophila)
|
|
chr3_-_31998241
|
0.413
|
NM_001174060
NM_017784
|
OSBPL10
|
oxysterol binding protein-like 10
|
|
chr3_-_126257425
|
0.413
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr19_-_18913011
|
0.413
|
|
HOMER3
|
homer homolog 3 (Drosophila)
|
|
chr17_+_45858517
|
0.412
|
NM_025149
|
ACSF2
|
acyl-CoA synthetase family member 2
|
|
chr3_-_31997738
|
0.411
|
|
OSBPL10
|
oxysterol binding protein-like 10
|
|
chr2_+_238265617
|
0.408
|
|
LRRFIP1
|
leucine rich repeat (in FLII) interacting protein 1
|
|
chr7_+_55054416
|
0.408
|
|
EGFR
|
epidermal growth factor receptor
|
|
chr20_+_336693
|
0.408
|
NM_006462
NM_031229
|
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1
|
|
chr20_+_25176698
|
0.407
|
NM_002862
|
PYGB
|
phosphorylase, glycogen; brain
|
|
chr13_+_97593469
|
0.406
|
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr16_-_4106186
|
0.406
|
NM_001116
|
ADCY9
|
adenylate cyclase 9
|
|
chr22_-_49292995
|
0.405
|
NM_033200
|
LMF2
|
lipase maturation factor 2
|
|
chr6_+_89847183
|
0.405
|
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr12_+_130945215
|
0.405
|
NM_003565
|
ULK1
|
unc-51-like kinase 1 (C. elegans)
|
|
chr17_+_50697355
|
0.404
|
|
HLF
|
hepatic leukemia factor
|
|
chr2_-_165186232
|
0.402
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr22_-_49292886
|
0.401
|
|
LMF2
|
lipase maturation factor 2
|
|
chr19_+_13736345
|
0.401
|
|
MRI1
|
methylthioribose-1-phosphate isomerase homolog (S. cerevisiae)
|
|
chr11_+_129445017
|
0.400
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr22_-_41375323
|
0.400
|
NM_000398
|
CYB5R3
|
cytochrome b5 reductase 3
|
|
chr16_-_55876099
|
0.399
|
|
PLLP
|
plasmolipin
|
|
chr11_+_118544625
|
0.399
|
NM_024618
NM_170722
|
NLRX1
|
NLR family member X1
|
|
chr17_+_50697319
|
0.398
|
NM_002126
|
HLF
|
hepatic leukemia factor
|
|
chr6_+_35418377
|
0.398
|
|
PPARD
|
peroxisome proliferator-activated receptor delta
|
|
chr8_+_86563382
|
0.395
|
NM_000067
|
CA2
|
carbonic anhydrase II
|
|
chr19_-_242168
|
0.393
|
NM_177543
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chr19_+_3045266
|
0.392
|
NM_002067
|
GNA11
|
guanine nucleotide binding protein (G protein), alpha 11 (Gq class)
|
|
chr8_+_136539290
|
0.391
|
|
KHDRBS3
|
KH domain containing, RNA binding, signal transduction associated 3
|
|
chr11_+_129445002
|
0.390
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr6_-_134538526
|
0.388
|
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr19_+_43447262
|
0.386
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr17_-_72218661
|
0.386
|
|
MXRA7
|
matrix-remodelling associated 7
|
|
chr8_+_26296439
|
0.386
|
NM_004331
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like
|
|
chr10_-_3817205
|
0.386
|
|
KLF6
|
Kruppel-like factor 6
|
|
chr2_+_173000552
|
0.385
|
NM_000210
NM_001079818
|
ITGA6
|
integrin, alpha 6
|
|
chr13_+_97593411
|
0.384
|
NM_001001715
NM_005766
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr3_-_187025501
|
0.384
|
NM_001007225
NM_006548
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr11_-_27450808
|
0.383
|
NM_018490
|
LGR4
|
leucine-rich repeat containing G protein-coupled receptor 4
|