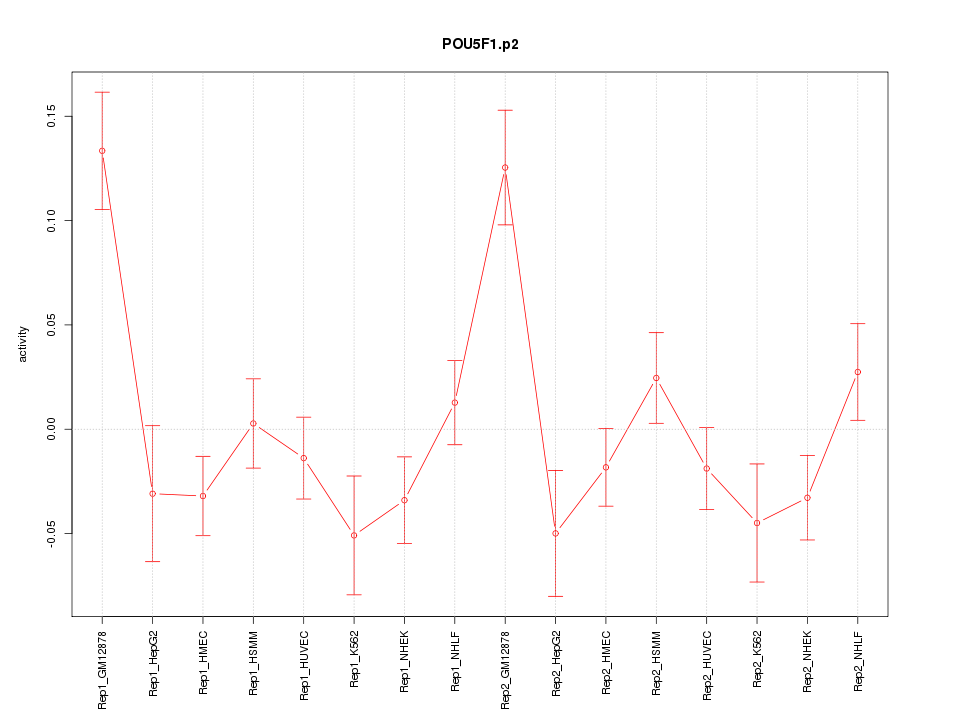
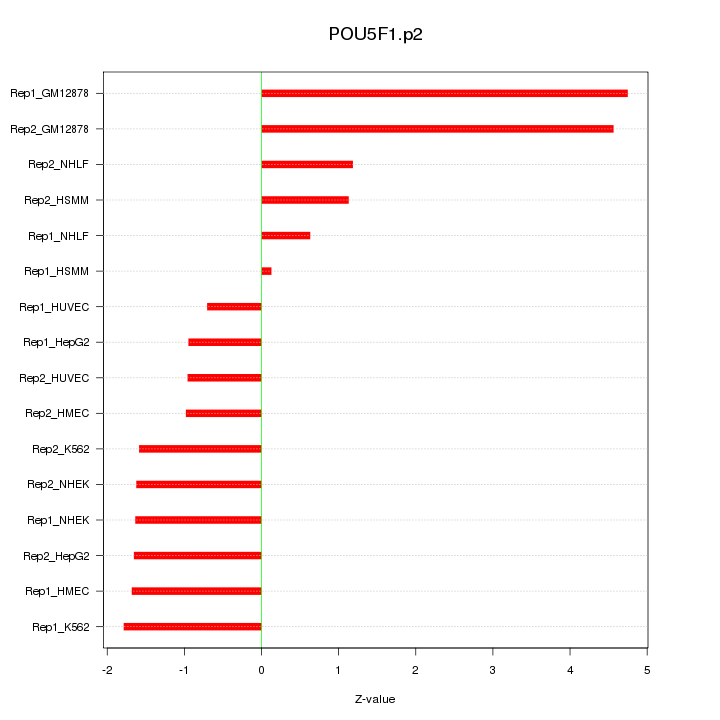
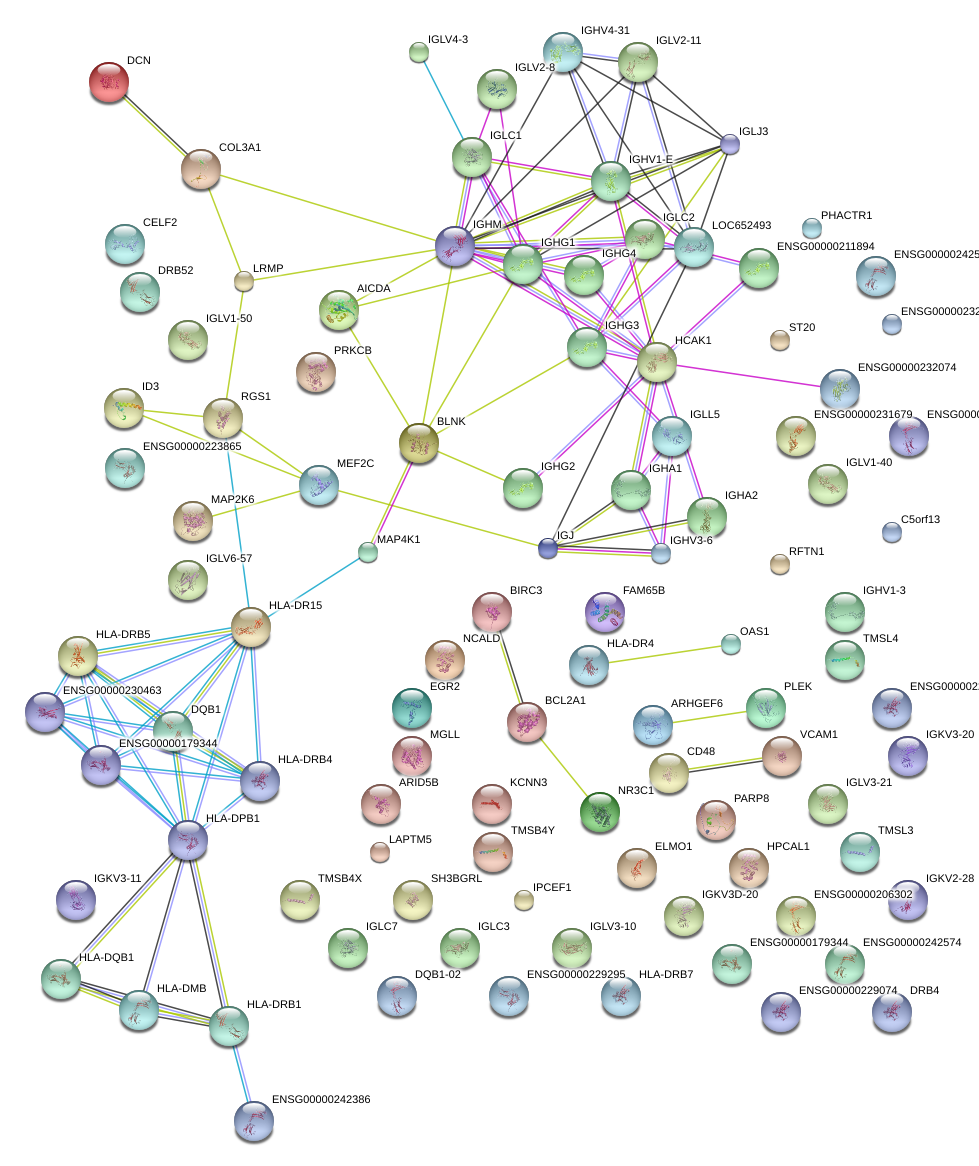
|
chr17_+_64922420
|
10.862
|
NM_002758
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr10_+_11246934
|
6.958
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr10_-_98021144
|
6.916
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr4_-_91979113
|
5.411
|
NM_183049
|
TMSL3
|
thymosin-like 3
|
|
chr22_+_21027538
|
4.942
|
|
|
|
|
chr22_+_21543671
|
4.814
|
|
IGLV4-3
|
immunoglobulin lambda variable 4-3
|
|
chr22_+_21116283
|
4.540
|
|
|
|
|
chr15_-_78050515
|
4.410
|
NM_001114735
NM_004049
|
BCL2A1
|
BCL2-related protein A1
|
|
chr1_-_42738535
|
4.408
|
|
TMSB4XP1
|
thymosin beta 4, X-linked pseudogene 1
|
|
chr22_+_21340757
|
4.163
|
|
|
|
|
chr22_+_21370394
|
4.073
|
|
IGL@
|
immunoglobulin lambda locus
|
|
chr14_-_106249999
|
4.070
|
|
IGHA1
|
immunoglobulin heavy constant alpha 1
|
|
chr14_-_105476678
|
3.881
|
|
IGHM
|
immunoglobulin heavy constant mu
|
|
chr6_+_12825022
|
3.841
|
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr2_+_88966170
|
3.824
|
|
|
|
|
chr22_+_21065134
|
3.818
|
|
LOC100290481
|
immunoglobulin lambda light chain-like
|
|
chr22_+_21464980
|
3.785
|
|
|
|
|
chr14_-_105997732
|
3.692
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr10_+_11247059
|
3.618
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr4_-_71751070
|
3.571
|
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides
|
|
chr22_+_21053972
|
3.570
|
|
|
|
|
chr19_-_43800374
|
3.525
|
NM_001042600
NM_007181
|
MAP4K1
|
mitogen-activated protein kinase kinase kinase kinase 1
|
|
chr7_-_36991189
|
3.505
|
NM_001039459
|
ELMO1
|
engulfment and cell motility 1
|
|
chr16_+_23754822
|
3.486
|
|
PRKCB
|
protein kinase C, beta
|
|
chr22_+_21495273
|
3.455
|
|
|
|
|
chr16_+_23754772
|
3.377
|
NM_002738
NM_212535
|
PRKCB
|
protein kinase C, beta
|
|
chr16_+_23754860
|
3.356
|
|
PRKCB
|
protein kinase C, beta
|
|
chr22_+_21407066
|
3.259
|
|
|
|
|
chr14_-_106270497
|
3.197
|
|
IGHG3
|
immunoglobulin heavy constant gamma 3 (G3m marker)
|
|
chr22_+_21065207
|
3.134
|
|
IGLJ3
|
immunoglobulin lambda joining 3
|
|
chr14_-_106184988
|
3.043
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr2_-_89223666
|
2.967
|
|
IGKC
IGK@
|
immunoglobulin kappa constant
immunoglobulin kappa locus
|
|
chr5_-_111119846
|
2.944
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr14_-_105804211
|
2.911
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr22_+_20880112
|
2.893
|
|
|
|
|
chr14_-_106084519
|
2.869
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr5_-_88155360
|
2.831
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr6_-_154692906
|
2.754
|
NM_001130699
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1
|
|
chr6_-_32665409
|
2.512
|
NM_002124
|
HLA-DRB1
HLA-DRB4
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 1
major histocompatibility complex, class II, DR beta 4
major histocompatibility complex, class II, DR beta 5
|
|
chr3_-_129023850
|
2.494
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr14_-_105762742
|
2.455
|
|
|
|
|
chr1_-_31003260
|
2.436
|
NM_006762
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chrX_+_80344215
|
2.406
|
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chr6_-_33016561
|
2.293
|
NM_002118
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta
|
|
chr1_-_31003181
|
2.255
|
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chrX_+_80343958
|
2.187
|
NM_003022
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chr14_-_106241445
|
2.180
|
|
IGHV4-31
IGHV1-69
IGHA1
IGHG1
IGHG3
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy variable 1-69
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
|
|
chr1_-_158948176
|
2.170
|
NM_001778
|
CD48
|
CD48 molecule
|
|
chr10_-_64248932
|
2.100
|
NM_001136178
|
EGR2
|
early growth response 2
|
|
chrX_+_80344253
|
2.091
|
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chr14_-_105681377
|
2.052
|
|
IGHV4-31
IGHG1
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr14_-_105644300
|
2.007
|
|
IGHV4-31
IGHA1
IGHA2
IGHG1
IGHG3
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
|
|
chr8_-_102872372
|
2.003
|
|
NCALD
|
neurocalcin delta
|
|
chr12_+_25096472
|
2.002
|
NM_006152
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr11_+_101693403
|
1.951
|
NM_001165
NM_182962
|
BIRC3
|
baculoviral IAP repeat containing 3
|
|
chr14_-_106149623
|
1.947
|
|
|
|
|
chr1_-_153098811
|
1.939
|
NM_170782
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr10_-_64246080
|
1.922
|
|
EGR2
|
early growth response 2
|
|
chr12_-_90100710
|
1.909
|
|
DCN
|
decorin
|
|
chr12_-_90100679
|
1.896
|
|
DCN
|
decorin
|
|
chr5_-_88215034
|
1.894
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr22_+_20880159
|
1.893
|
|
IGLV6-57
|
immunoglobulin lambda variable 6-57
|
|
chr12_+_111829121
|
1.880
|
NM_001032409
NM_002534
NM_016816
|
OAS1
|
2',5'-oligoadenylate synthetase 1, 40/46kDa
|
|
chrX_-_135690770
|
1.868
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chrX_-_135690699
|
1.860
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr10_-_64246129
|
1.859
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr5_-_142764168
|
1.849
|
NM_001018076
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr2_+_68445821
|
1.842
|
NM_002664
|
PLEK
|
pleckstrin
|
|
chr12_-_8656705
|
1.824
|
NM_020661
|
AICDA
|
activation-induced cytidine deaminase
|
|
chr1_-_23758503
|
1.817
|
|
ID3
|
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein
|
|
chr5_+_49998677
|
1.799
|
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr1_+_100957883
|
1.794
|
NM_001078
NM_080682
|
VCAM1
|
vascular cell adhesion molecule 1
|
|
chr5_+_49998528
|
1.793
|
NM_001178056
NM_024615
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr3_-_16530132
|
1.745
|
NM_015150
|
RFTN1
|
raftlin, lipid raft linker 1
|
|
chr14_-_105589467
|
1.744
|
|
LOC100126583
IGHA1
IGHG1
IGHG2
IGHM
|
hypothetical LOC100126583
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 2 (G2m marker)
immunoglobulin heavy constant mu
|
|
chr14_-_105712630
|
1.742
|
|
IGHV4-31
IGHA1
IGHA2
IGHG1
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr2_+_189547377
|
1.740
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr12_-_90100733
|
1.694
|
NM_001920
|
DCN
|
decorin
|
|
chr12_+_25096755
|
1.686
|
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr22_+_21065202
|
1.670
|
|
|
|
|
chr1_+_190811479
|
1.665
|
NM_002922
|
RGS1
|
regulator of G-protein signaling 1
|
|
chr14_-_106193403
|
1.658
|
|
IGKV3-20
|
immunoglobulin kappa variable 3-20
|
|
chr2_+_189547358
|
1.652
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr6_-_24985432
|
1.643
|
NM_015864
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chr10_+_63331018
|
1.639
|
NM_032199
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr18_-_51406359
|
1.633
|
|
TCF4
|
transcription factor 4
|
|
chr12_+_14409875
|
1.624
|
NM_018179
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr13_-_98757634
|
1.616
|
NM_004951
|
GPR183
|
G protein-coupled receptor 183
|
|
chr18_-_51406841
|
1.614
|
NM_001083962
NM_003199
|
TCF4
|
transcription factor 4
|
|
chr17_-_59363352
|
1.605
|
NM_000626
NM_001039933
NM_021602
|
CD79B
|
CD79b molecule, immunoglobulin-associated beta
|
|
chr22_+_21042091
|
1.603
|
|
IGLV1-44
IGL@
|
immunoglobulin lambda variable 1-44
immunoglobulin lambda locus
|
|
chr5_+_49997489
|
1.542
|
NM_001178055
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr17_-_38087366
|
1.538
|
NM_016602
|
CCR10
|
chemokine (C-C motif) receptor 10
|
|
chr4_+_68107039
|
1.529
|
NM_012108
|
STAP1
|
signal transducing adaptor family member 1
|
|
chr8_-_102872354
|
1.520
|
|
NCALD
|
neurocalcin delta
|
|
chr14_+_63752424
|
1.508
|
NM_182910
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2
|
|
chr14_-_106064887
|
1.506
|
|
LOC100126583
IGHV3-48
IGHA1
IGHM
|
hypothetical LOC100126583
immunoglobulin heavy variable 3-48
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant mu
|
|
chr3_+_99733432
|
1.490
|
NM_005290
|
GPR15
|
G protein-coupled receptor 15
|
|
chrX_-_135691168
|
1.488
|
NM_004840
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr14_-_105565211
|
1.486
|
|
IGHV4-31
IGHA1
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
|
|
chr3_-_126325132
|
1.442
|
|
SLC12A8
|
solute carrier family 12 (potassium/chloride transporters), member 8
|
|
chrX_+_12903145
|
1.414
|
NM_021109
NM_183049
|
TMSB4X
TMSL3
|
thymosin beta 4, X-linked
thymosin-like 3
|
|
chrX_-_106846924
|
1.386
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr6_-_32892705
|
1.378
|
NM_002120
|
HLA-DOB
|
major histocompatibility complex, class II, DO beta
|
|
chr21_+_25933916
|
1.373
|
|
JAM2
|
junctional adhesion molecule 2
|
|
chr8_+_56954953
|
1.362
|
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
|
|
chr2_+_89715037
|
1.347
|
|
|
|
|
chr5_-_111120764
|
1.326
|
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chrX_-_70248022
|
1.325
|
NM_000206
|
IL2RG
|
interleukin 2 receptor, gamma
|
|
chr5_-_111340421
|
1.325
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr3_+_88270951
|
1.323
|
NM_018293
|
ZNF654
|
zinc finger protein 654
|
|
chr5_-_88235624
|
1.316
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr14_-_105763152
|
1.312
|
|
IGHV4-31
IGHA1
IGHD
IGHG1
IGHG3
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
|
|
chr1_-_111544784
|
1.312
|
NM_024901
|
DENND2D
|
DENN/MADD domain containing 2D
|
|
chr12_-_101398451
|
1.298
|
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr21_+_25933938
|
1.297
|
|
JAM2
|
junctional adhesion molecule 2
|
|
chr3_+_179736917
|
1.286
|
NM_181361
|
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2
|
|
chr8_+_56954899
|
1.277
|
NM_001111097
NM_002350
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
|
|
chrX_-_106846684
|
1.272
|
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr2_+_189547323
|
1.263
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr7_+_73826244
|
1.233
|
NM_000265
|
NCF1
NCF1B
NCF1C
|
neutrophil cytosolic factor 1
neutrophil cytosolic factor 1B pseudogene
neutrophil cytosolic factor 1C pseudogene
|
|
chr3_-_58175437
|
1.231
|
NM_004944
|
DNASE1L3
|
deoxyribonuclease I-like 3
|
|
chr8_+_56954946
|
1.230
|
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
|
|
chr5_-_111120817
|
1.225
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chrX_+_154264931
|
1.215
|
NM_001007523
NM_001007524
NM_012151
|
F8A2
F8A3
F8A1
|
coagulation factor VIII-associated (intronic transcript) 2
coagulation factor VIII-associated (intronic transcript) 3
coagulation factor VIII-associated (intronic transcript) 1
|
|
chr6_-_137582237
|
1.204
|
|
IFNGR1
|
interferon gamma receptor 1
|
|
chrX_-_153767625
|
1.200
|
NM_019863
|
F8
|
coagulation factor VIII, procoagulant component
|
|
chr4_-_153493133
|
1.185
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr2_-_191580567
|
1.179
|
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa
|
|
chr2_+_33514896
|
1.177
|
NM_170672
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr4_+_40953652
|
1.174
|
NM_004181
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr22_+_21393107
|
1.172
|
|
IGLJ3
|
immunoglobulin lambda joining 3
|
|
chr6_+_32515596
|
1.167
|
NM_019111
|
HLA-DRA
|
major histocompatibility complex, class II, DR alpha
|
|
chr1_-_111544820
|
1.162
|
|
DENND2D
|
DENN/MADD domain containing 2D
|
|
chr6_-_137582244
|
1.150
|
NM_000416
|
IFNGR1
|
interferon gamma receptor 1
|
|
chr11_+_128068982
|
1.134
|
NM_002017
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr17_-_45617899
|
1.126
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chrX_-_80344096
|
1.125
|
NM_030763
|
HMGN5
|
high-mobility group nucleosome binding domain 5
|
|
chr3_-_16529996
|
1.124
|
|
RFTN1
|
raftlin, lipid raft linker 1
|
|
chr22_+_21094094
|
1.118
|
|
|
|
|
chr6_-_133120587
|
1.112
|
NM_004665
|
VNN2
|
vanin 2
|
|
chr14_-_105623358
|
1.096
|
|
IGHA1
IGHD
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr22_-_22426551
|
1.095
|
NM_013378
|
VPREB3
|
pre-B lymphocyte 3
|
|
chr7_+_116447527
|
1.095
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr6_-_29635456
|
1.093
|
|
UBD
|
ubiquitin D
|
|
chr14_-_106119742
|
1.089
|
|
IGLJ3
IGHG1
|
immunoglobulin lambda joining 3
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr4_-_176971175
|
1.087
|
NM_201591
|
GPM6A
|
glycoprotein M6A
|
|
chr15_-_43458247
|
1.070
|
NM_001482
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase)
|
|
chr10_+_91051685
|
1.065
|
NM_001547
|
IFIT2
|
interferon-induced protein with tetratricopeptide repeats 2
|
|
chr12_-_101398503
|
1.065
|
NM_000618
NM_001111283
NM_001111285
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr2_+_237143118
|
1.062
|
NM_020311
|
CXCR7
|
chemokine (C-X-C motif) receptor 7
|
|
chr17_-_39094787
|
1.060
|
NM_001040002
|
MEOX1
|
mesenchyme homeobox 1
|
|
chr4_+_40953666
|
1.053
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr12_-_101398429
|
1.052
|
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chrX_+_153767803
|
1.050
|
NM_001007523
NM_001007524
NM_012151
|
F8A2
F8A3
F8A1
|
coagulation factor VIII-associated (intronic transcript) 2
coagulation factor VIII-associated (intronic transcript) 3
coagulation factor VIII-associated (intronic transcript) 1
|
|
chr2_-_89400935
|
1.039
|
|
IGKC
|
immunoglobulin kappa constant
|
|
chr3_+_135996734
|
1.037
|
NM_004441
|
EPHB1
|
EPH receptor B1
|
|
chr19_+_40511909
|
1.027
|
NM_001185099
NM_001185100
NM_001185101
NM_001771
|
CD22
|
CD22 molecule
|
|
chr13_-_98708549
|
1.025
|
NM_001098200
NM_005292
|
GPR18
|
G protein-coupled receptor 18
|
|
chr4_-_71751205
|
1.024
|
NM_144646
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides
|
|
chr2_-_89294466
|
1.023
|
|
IGKC
|
immunoglobulin kappa constant
|
|
chr3_-_57209319
|
1.022
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chr1_+_154386358
|
1.017
|
NM_001193301
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr14_-_22354851
|
1.006
|
NM_001126105
NM_003982
|
SLC7A7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7
|
|
chr1_+_208008517
|
1.005
|
|
TRAF3IP3
|
TRAF3 interacting protein 3
|
|
chrX_-_154341451
|
1.000
|
NM_001007523
NM_001007524
NM_012151
|
F8A2
F8A3
F8A1
|
coagulation factor VIII-associated (intronic transcript) 2
coagulation factor VIII-associated (intronic transcript) 3
coagulation factor VIII-associated (intronic transcript) 1
|
|
chr16_+_30391460
|
0.990
|
NM_001114380
NM_002209
|
ITGAL
|
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide)
|
|
chrX_-_33267646
|
0.987
|
NM_000109
|
DMD
|
dystrophin
|
|
chr2_+_73973600
|
0.986
|
NM_001615
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr4_-_176970895
|
0.976
|
|
GPM6A
|
glycoprotein M6A
|
|
chr2_+_73973640
|
0.967
|
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr8_+_70567634
|
0.958
|
|
SULF1
|
sulfatase 1
|
|
chr8_+_26569709
|
0.953
|
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr12_+_111829191
|
0.953
|
|
OAS1
|
2',5'-oligoadenylate synthetase 1, 40/46kDa
|
|
chr5_-_95184221
|
0.950
|
NM_001118890
NM_002064
|
GLRX
|
glutaredoxin (thioltransferase)
|
|
chr1_+_63861474
|
0.949
|
NM_001172818
|
PGM1
|
phosphoglucomutase 1
|
|
chr12_-_90029328
|
0.925
|
|
LUM
|
lumican
|
|
chr12_+_27288344
|
0.923
|
NM_015000
|
STK38L
|
serine/threonine kinase 38 like
|
|
chrX_-_62891066
|
0.918
|
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr14_-_106170414
|
0.913
|
|
IGHG1
IGHG3
|
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
|
|
chr11_-_58099884
|
0.907
|
NM_004811
|
LPXN
|
leupaxin
|
|
chr13_-_40491405
|
0.905
|
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr3_-_126412882
|
0.896
|
NM_001195483
|
SLC12A8
|
solute carrier family 12 (potassium/chloride transporters), member 8
|
|
chr1_+_242281175
|
0.895
|
NM_205768
|
ZNF238
|
zinc finger protein 238
|
|
chrX_-_48661226
|
0.892
|
NM_006875
|
PIM2
|
pim-2 oncogene
|
|
chr4_+_41395493
|
0.888
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr22_+_20715340
|
0.885
|
|
|
|
|
chr10_-_91001566
|
0.882
|
NM_000235
NM_001127605
|
LIPA
|
lipase A, lysosomal acid, cholesterol esterase
|
|
chr11_+_128069183
|
0.880
|
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr1_+_154120170
|
0.872
|
|
SYT11
|
synaptotagmin XI
|
|
chr7_+_99654806
|
0.852
|
NM_024070
|
PVRIG
|
poliovirus receptor related immunoglobulin domain containing
|
|
chr14_-_105851580
|
0.847
|
|
IGHA1
IGHA2
IGHD
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr10_-_91001496
|
0.846
|
|
LIPA
|
lipase A, lysosomal acid, cholesterol esterase
|
|
chr12_-_7136189
|
0.844
|
|
C1R
|
complement component 1, r subcomponent
|
|
chr8_+_26491317
|
0.844
|
NM_001386
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr4_+_40953698
|
0.844
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr14_-_105610144
|
0.843
|
|
IGHA1
IGHG1
IGHG3
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
|
|
chr12_-_101398288
|
0.842
|
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr4_-_176970407
|
0.829
|
|
GPM6A
|
glycoprotein M6A
|
|
chr15_-_91078307
|
0.828
|
|
FAM174B
|
family with sequence similarity 174, member B
|