|
chr8_-_49996353
|
3.595
|
NM_003068
|
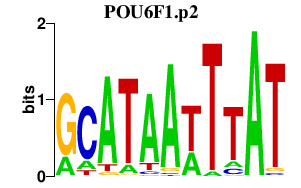
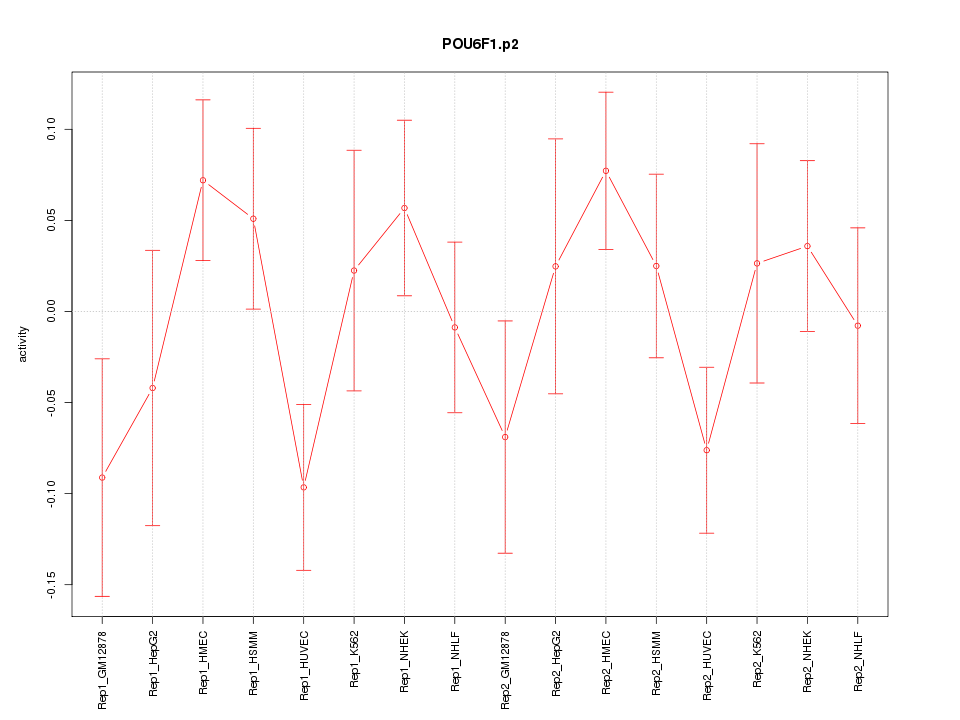
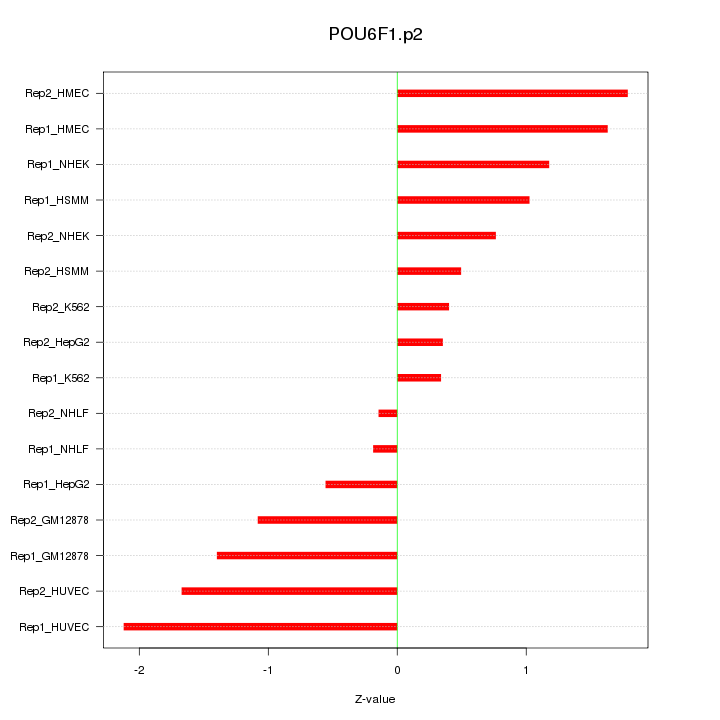
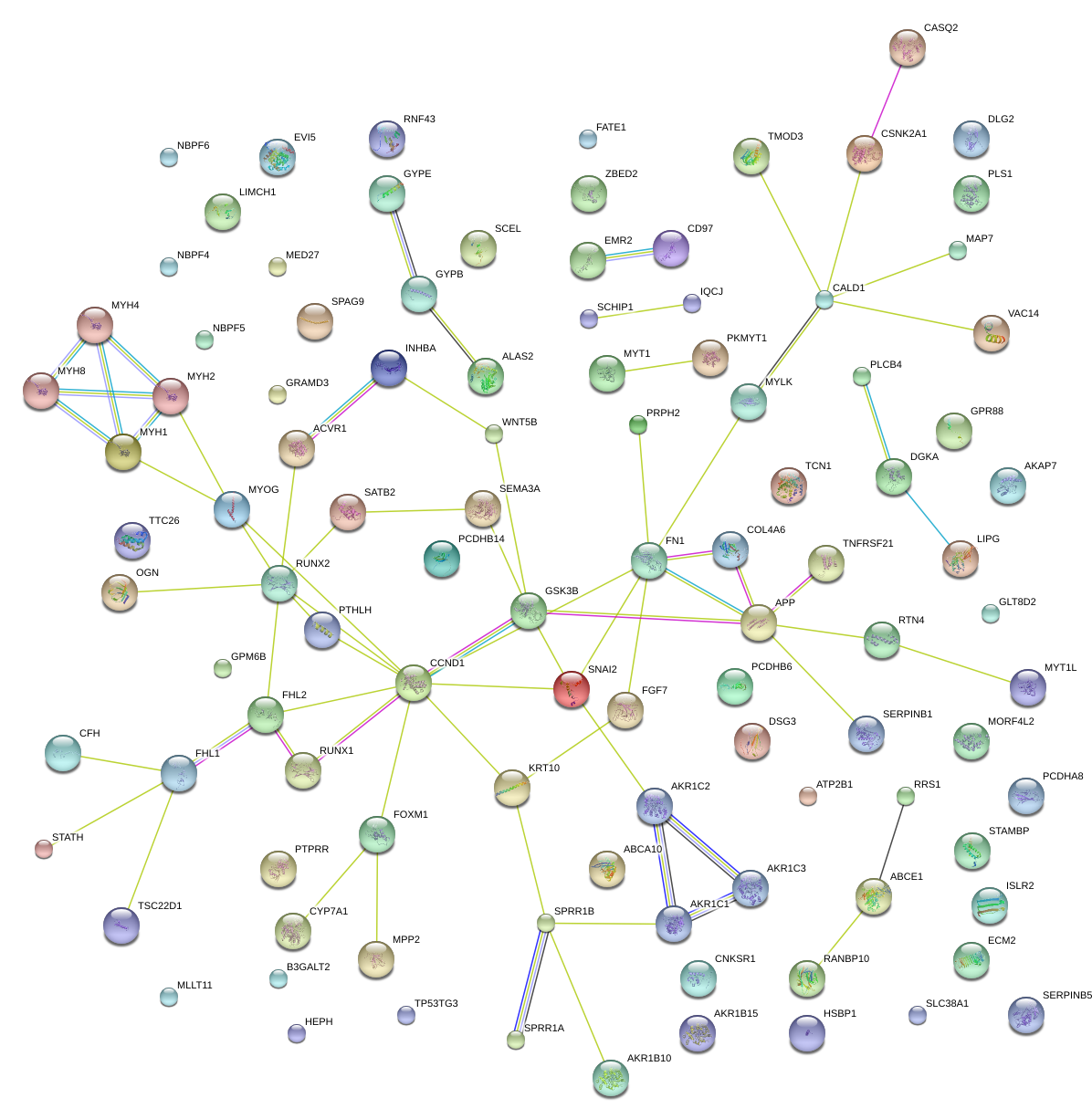
SNAI2
|
snail homolog 2 (Drosophila)
|
|
chr5_+_125786717
|
2.882
|
NM_001146320
NM_001146321
NM_023927
|
GRAMD3
|
GRAM domain containing 3
|
|
chr1_+_151270301
|
2.560
|
NM_003125
|
SPRR1B
|
small proline-rich protein 1B
|
|
chr5_+_125786975
|
2.530
|
|
GRAMD3
|
GRAM domain containing 3
|
|
chr10_+_4995685
|
2.297
|
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr10_+_4995453
|
2.191
|
NM_001353
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr18_+_59295123
|
1.964
|
NM_002639
|
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5
|
|
chr3_-_112796855
|
1.752
|
NM_024508
|
ZBED2
|
zinc finger, BED-type containing 2
|
|
chr11_+_69176391
|
1.643
|
|
CCND1
|
cyclin D1
|
|
chr13_-_43908994
|
1.545
|
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr13_-_43908985
|
1.467
|
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr13_-_43908937
|
1.462
|
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr10_+_4995597
|
1.458
|
|
AKR1C1
AKR1C3
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr13_-_43908995
|
1.412
|
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr13_-_43908972
|
1.331
|
NM_006022
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr10_-_5035967
|
1.277
|
NM_001135241
|
AKR1C1
AKR1C2
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III)
|
|
chr12_-_28016182
|
1.061
|
NM_002820
NM_198965
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr6_-_47362102
|
1.023
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chrX_+_65299157
|
0.997
|
NM_138737
|
HEPH
|
hephaestin
|
|
chr1_-_116112853
|
0.842
|
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr1_-_116112921
|
0.829
|
NM_001232
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr1_-_191422346
|
0.829
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr1_-_116112684
|
0.770
|
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr3_+_143858428
|
0.770
|
NM_001172312
|
PLS1
|
plastin 1
|
|
chrX_+_135058402
|
0.715
|
NM_001159700
|
FHL1
|
four and a half LIM domains 1
|
|
chr2_-_215949566
|
0.694
|
|
FN1
|
fibronectin 1
|
|
chrX_+_135106578
|
0.682
|
NM_001159701
NM_001159699
|
FHL1
|
four and a half LIM domains 1
|
|
chr6_-_136888771
|
0.672
|
NM_001198608
NM_001198609
NM_001198611
|
MAP7
|
microtubule-associated protein 7
|
|
chr1_+_15145001
|
0.649
|
NM_001017999
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr6_-_47361921
|
0.624
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr3_-_124821868
|
0.610
|
|
MYLK
|
myosin light chain kinase
|
|
chr12_+_54611212
|
0.600
|
NM_001345
|
DGKA
|
diacylglycerol kinase, alpha 80kDa
|
|
chr22_-_34566276
|
0.580
|
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr22_-_34566367
|
0.579
|
NM_001031695
NM_001082576
NM_001082577
NM_014309
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chrX_-_13744933
|
0.577
|
|
GPM6B
|
glycoprotein M6B
|
|
chr2_-_158439868
|
0.557
|
NM_001105
|
ACVR1
|
activin A receptor, type I
|
|
chr22_-_34566210
|
0.545
|
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chrX_-_107569256
|
0.539
|
NM_001847
|
COL4A6
|
collagen, type IV, alpha 6
|
|
chr5_+_125723686
|
0.522
|
NM_001146319
|
GRAMD3
|
GRAM domain containing 3
|
|
chr18_+_27281729
|
0.505
|
NM_001944
|
DSG3
|
desmoglein 3
|
|
chr17_-_46479127
|
0.502
|
|
SPAG9
|
sperm associated antigen 9
|
|
chr4_-_145281237
|
0.497
|
NM_002099
|
GYPA
|
glycophorin A (MNS blood group)
|
|
chr7_+_133862883
|
0.487
|
NM_020299
|
AKR1B10
|
aldo-keto reductase family 1, member B10 (aldose reductase)
|
|
chr17_-_10265991
|
0.482
|
NM_002472
|
MYH8
|
myosin, heavy chain 8, skeletal muscle, perinatal
|
|
chr12_-_88573949
|
0.466
|
NM_001001323
NM_001682
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1
|
|
chr11_-_59390518
|
0.424
|
NM_001062
|
TCN1
|
transcobalamin I (vitamin B12 binding protein, R binder family)
|
|
chr6_-_47385470
|
0.419
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chrX_+_146941222
|
0.386
|
|
FTH1P8
|
ferritin, heavy polypeptide 1 pseudogene 8
|
|
chrX_+_65299352
|
0.371
|
|
HEPH
|
hephaestin
|
|
chr9_-_133944877
|
0.370
|
|
MED27
|
mediator complex subunit 27
|
|
chr20_+_9146036
|
0.355
|
NM_182797
|
PLCB4
|
phospholipase C, beta 4
|
|
chr1_+_194887630
|
0.328
|
NM_000186
NM_001014975
|
CFH
|
complement factor H
|
|
chr11_-_11331411
|
0.318
|
|
CSNK2A1P
|
casein kinase 2, alpha 1 polypeptide pseudogene
|
|
chr13_+_77007809
|
0.312
|
NM_001160706
NM_003843
NM_144777
|
SCEL
|
sciellin
|
|
chr17_-_36232366
|
0.305
|
NM_000421
|
KRT10
|
keratin 10
|
|
chr9_-_94206648
|
0.302
|
NM_014057
NM_033014
|
OGN
|
osteoglycin
|
|
chr6_-_2785682
|
0.299
|
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1
|
|
chr7_+_134114912
|
0.285
|
|
CALD1
|
caldesmon 1
|
|
chr6_-_47385605
|
0.284
|
NM_014452
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr3_-_124822113
|
0.279
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr3_+_160269734
|
0.279
|
NM_001197113
NM_001197114
NM_001042705
NM_001042706
NM_001197100
|
IQCJ-SCHIP1
IQCJ
|
IQCJ-SCHIP1 readthrough
IQ motif containing J
|
|
chr18_+_45342398
|
0.279
|
NM_006033
|
LIPG
|
lipase, endothelial
|
|
chr2_-_1905508
|
0.278
|
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr16_-_69392568
|
0.275
|
|
VAC14
|
Vac14 homolog (S. cerevisiae)
|
|
chr7_+_134114957
|
0.270
|
|
CALD1
|
caldesmon 1
|
|
chrX_+_150635162
|
0.270
|
NM_033085
|
FATE1
|
fetal and adult testis expressed 1
|
|
chr8_+_67505091
|
0.267
|
|
RRS1
|
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae)
|
|
chr16_-_69392561
|
0.266
|
NM_018052
|
VAC14
|
Vac14 homolog (S. cerevisiae)
|
|
chr15_+_49988882
|
0.266
|
|
TMOD3
|
tropomodulin 3 (ubiquitous)
|
|
chr9_-_94338072
|
0.260
|
NM_001197295
NM_001197296
NM_001393
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific
|
|
chr15_+_72209795
|
0.258
|
NM_020851
|
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2
|
|
chr15_+_72209973
|
0.255
|
|
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2
|
|
chr7_-_83662152
|
0.255
|
NM_006080
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A
|
|
chr21_-_35343456
|
0.255
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr1_+_26376560
|
0.255
|
NM_006314
|
CNKSR1
|
connector enhancer of kinase suppressor of Ras 1
|
|
chr2_+_73911477
|
0.254
|
|
STAMBP
|
STAM binding protein
|
|
chr12_-_44919818
|
0.253
|
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr6_+_131508153
|
0.252
|
NM_016377
|
AKAP7
|
A kinase (PRKA) anchor protein 7
|
|
chr16_-_69392562
|
0.251
|
|
VAC14
|
Vac14 homolog (S. cerevisiae)
|
|
chr12_-_69434330
|
0.250
|
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr5_+_140200995
|
0.245
|
NM_018911
NM_031856
|
PCDHA8
|
protocadherin alpha 8
|
|
chr17_-_53847937
|
0.230
|
|
RNF43
|
ring finger protein 43
|
|
chr7_+_138469029
|
0.222
|
NM_001144920
NM_001144923
NM_024926
|
TTC26
|
tetratricopeptide repeat domain 26
|
|
chr7_-_41709191
|
0.221
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr15_+_47502666
|
0.215
|
NM_002009
|
FGF7
|
fibroblast growth factor 7
|
|
chr4_-_145159889
|
0.214
|
NM_002100
|
GYPB
|
glycophorin B (MNS blood group)
|
|
chr1_-_108588186
|
0.209
|
NM_001143989
|
NBPF4
NBPF6
|
neuroblastoma breakpoint family, member 4
neuroblastoma breakpoint family, member 6
|
|
chr19_-_14750347
|
0.206
|
NM_013447
NM_152916
NM_152917
NM_152918
NM_152919
NM_152920
NM_152921
|
EMR2
|
egf-like module containing, mucin-like, hormone receptor-like 2
|
|
chr3_-_121117647
|
0.200
|
|
GSK3B
|
glycogen synthase kinase 3 beta
|
|
chr4_+_41309668
|
0.199
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr4_+_146250978
|
0.198
|
|
ABCE1
|
ATP-binding cassette, sub-family E (OABP), member 1
|
|
chr21_-_35343500
|
0.195
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chrX_-_102828238
|
0.194
|
NM_001142418
NM_001142423
NM_001142424
NM_001142425
NM_001142426
NM_001142429
NM_001142430
NM_012286
|
MORF4L2
|
mortality factor 4 like 2
|
|
chr1_+_149299307
|
0.192
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr1_+_100776280
|
0.191
|
NM_022049
|
GPR88
|
G protein-coupled receptor 88
|
|
chrX_-_55074125
|
0.188
|
NM_000032
NM_001037967
NM_001037968
|
ALAS2
|
aminolevulinate, delta-, synthase 2
|
|
chr1_-_201321711
|
0.184
|
|
MYOG
|
myogenin (myogenic factor 4)
|
|
chr1_-_93030538
|
0.178
|
NM_005665
|
EVI5
|
ecotropic viral integration site 5
|
|
chr2_-_55090759
|
0.174
|
NM_007008
|
RTN4
|
reticulon 4
|
|
chr2_-_200033445
|
0.174
|
|
SATB2
|
SATB homeobox 2
|
|
chr21_-_26269298
|
0.170
|
|
APP
|
amyloid beta (A4) precursor protein
|
|
chr8_-_59575249
|
0.169
|
NM_000780
|
CYP7A1
|
cytochrome P450, family 7, subfamily A, polypeptide 1
|
|
chr16_-_32595553
|
0.168
|
NM_016212
|
TP53TG3
|
TP53 target 3
|
|
chr4_+_70896236
|
0.167
|
NM_001009181
NM_003154
|
STATH
|
statherin
|
|
chr2_-_105421661
|
0.166
|
NM_201557
|
FHL2
|
four and a half LIM domains 2
|
|
chr12_+_1596482
|
0.164
|
NM_030775
|
WNT5B
|
wingless-type MMTV integration site family, member 5B
|
|
chr17_-_64752550
|
0.162
|
NM_080282
|
ABCA10
|
ATP-binding cassette, sub-family A (ABC1), member 10
|
|
chr5_+_140583098
|
0.160
|
NM_018934
|
PCDHB14
|
protocadherin beta 14
|
|
chr5_+_140509803
|
0.159
|
NM_018939
|
PCDHB6
|
protocadherin beta 6
|
|
chr16_-_69392385
|
0.158
|
|
VAC14
|
Vac14 homolog (S. cerevisiae)
|
|
chr12_-_69434650
|
0.157
|
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr12_-_102967828
|
0.154
|
|
GLT8D2
|
glycosyltransferase 8 domain containing 2
|
|
chr20_+_62266270
|
0.152
|
NM_004535
|
MYT1
|
myelin transcription factor 1
|
|
chr11_-_83071084
|
0.152
|
NM_001142702
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr3_-_139960874
|
0.152
|
NM_006219
|
PIK3CB
|
phosphoinositide-3-kinase, catalytic, beta polypeptide
|
|
chr16_+_33112480
|
0.150
|
NM_016212
|
TP53TG3
|
TP53 target 3
|
|
chr12_-_69434598
|
0.149
|
NM_130846
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr2_+_183290912
|
0.149
|
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10
|
|
chr16_-_69392545
|
0.147
|
|
VAC14
|
Vac14 homolog (S. cerevisiae)
|
|
chr8_+_32698892
|
0.146
|
NM_001159996
|
NRG1
|
neuregulin 1
|
|
chr3_+_129927714
|
0.143
|
|
RAB7A
|
RAB7A, member RAS oncogene family
|
|
chrX_-_153698930
|
0.141
|
|
HMGN1P37
|
high-mobility group nucleosome binding domain 1 pseudogene 37
|
|
chr1_+_213245507
|
0.137
|
NM_001017424
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr16_+_33169015
|
0.135
|
NM_016212
|
TP53TG3
|
TP53 target 3
|
|
chr17_+_46176105
|
0.134
|
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae)
|
|
chr7_+_120490054
|
0.133
|
|
C7orf58
|
chromosome 7 open reading frame 58
|
|
chr16_-_57309625
|
0.133
|
|
GOT2
|
glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2)
|
|
chrX_-_33139349
|
0.131
|
NM_004006
|
DMD
|
dystrophin
|
|
chr3_-_187309442
|
0.130
|
NM_004454
|
ETV5
|
ets variant 5
|
|
chr9_-_133945064
|
0.129
|
NM_004269
|
CRSP8P
MED27
|
mediator complex subunit 27 pseudogene
mediator complex subunit 27
|
|
chr21_-_35153664
|
0.129
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr4_-_145046109
|
0.128
|
NM_002102
NM_198682
|
GYPE
|
glycophorin E (MNS blood group)
|
|
chrX_+_123062143
|
0.128
|
|
STAG2
|
stromal antigen 2
|
|
chr3_+_129927716
|
0.128
|
|
RAB7A
|
RAB7A, member RAS oncogene family
|
|
chr11_+_112337199
|
0.126
|
NM_000615
NM_001076682
NM_181351
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr2_+_201806410
|
0.124
|
NM_001080124
NM_001228
NM_033355
NM_033358
|
CASP8
|
caspase 8, apoptosis-related cysteine peptidase
|
|
chr7_+_94135384
|
0.121
|
|
PEG10
|
paternally expressed 10
|
|
chr1_-_245120900
|
0.121
|
|
AHCTF1
|
AT hook containing transcription factor 1
|
|
chr17_-_36346240
|
0.121
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr9_+_21399132
|
0.121
|
NM_002170
|
IFNA8
|
interferon, alpha 8
|
|
chr10_+_102097246
|
0.120
|
|
|
|
|
chr3_+_190831909
|
0.118
|
NM_001114978
NM_001114979
NM_003722
|
TP63
|
tumor protein p63
|
|
chr7_+_93373755
|
0.118
|
NM_021955
|
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1
|
|
chr14_-_64479198
|
0.117
|
NM_002083
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal)
|
|
chr10_-_61819493
|
0.117
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chrX_-_52752935
|
0.116
|
NM_003147
NM_175698
NM_001164417
|
SSX2
SSX2B
|
synovial sarcoma, X breakpoint 2
synovial sarcoma, X breakpoint 2B
|
|
chr1_-_178100204
|
0.115
|
|
|
|
|
chr6_-_49942147
|
0.115
|
NM_001131
NM_170609
|
CRISP1
|
cysteine-rich secretory protein 1
|
|
chr3_+_70068440
|
0.115
|
NM_000248
NM_001184968
NM_198158
NM_198178
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr8_+_54926920
|
0.114
|
NM_170587
|
RGS20
|
regulator of G-protein signaling 20
|
|
chr6_+_152053323
|
0.112
|
NM_001122742
|
ESR1
|
estrogen receptor 1
|
|
chr7_-_50596238
|
0.110
|
NM_000790
|
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase)
|
|
chr7_-_63660879
|
0.110
|
|
|
|
|
chr4_+_95595419
|
0.109
|
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr15_+_38296920
|
0.109
|
NM_001128628
NM_001128629
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chrX_+_55760774
|
0.109
|
NM_006064
NM_016656
|
RRAGB
|
Ras-related GTP binding B
|
|
chr2_+_166034402
|
0.108
|
NM_001172173
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chrX_+_77889861
|
0.107
|
NM_005296
|
LPAR4
|
lysophosphatidic acid receptor 4
|
|
chr16_+_4842077
|
0.107
|
NM_016936
|
UBN1
|
ubinuclein 1
|
|
chr16_-_31347151
|
0.105
|
NM_005205
|
COX6A2
|
cytochrome c oxidase subunit VIa polypeptide 2
|
|
chr12_-_16652202
|
0.104
|
NM_001001395
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr10_-_29794484
|
0.104
|
|
SVIL
|
supervillin
|
|
chr1_+_149299490
|
0.103
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr4_+_107456215
|
0.102
|
NM_001142415
NM_004757
|
AIMP1
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 1
|
|
chrX_+_46935035
|
0.099
|
NM_153280
|
UBA1
|
ubiquitin-like modifier activating enzyme 1
|
|
chr6_+_155579788
|
0.099
|
NM_001010927
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2
|
|
chr3_+_129927723
|
0.098
|
|
RAB7A
|
RAB7A, member RAS oncogene family
|
|
chr6_+_29187565
|
0.097
|
NM_001005216
|
OR2J3
|
olfactory receptor, family 2, subfamily J, member 3
|
|
chr3_+_129927668
|
0.097
|
NM_004637
|
RAB7A
|
RAB7A, member RAS oncogene family
|
|
chr10_+_96433240
|
0.096
|
NM_000772
NM_001128925
|
CYP2C18
|
cytochrome P450, family 2, subfamily C, polypeptide 18
|
|
chr9_-_21177529
|
0.095
|
NM_021068
|
IFNA4
|
interferon, alpha 4
|
|
chr2_+_201464884
|
0.094
|
NM_001142356
|
NIF3L1
|
NIF3 NGG1 interacting factor 3-like 1 (S. pombe)
|
|
chr17_-_7159389
|
0.089
|
|
GPS2
|
G protein pathway suppressor 2
|
|
chr21_-_35343448
|
0.089
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr1_-_56750235
|
0.088
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr1_-_93172119
|
0.088
|
|
|
|
|
chr1_-_116184846
|
0.088
|
NM_001111061
|
NHLH2
|
nescient helix loop helix 2
|
|
chr12_-_47250069
|
0.088
|
NM_002289
|
LALBA
|
lactalbumin, alpha-
|
|
chr8_-_91164100
|
0.087
|
NM_004929
|
CALB1
|
calbindin 1, 28kDa
|
|
chr15_+_49909147
|
0.084
|
|
TMOD3
|
tropomodulin 3 (ubiquitous)
|
|
chr8_+_1980564
|
0.083
|
NM_003970
|
MYOM2
|
myomesin (M-protein) 2, 165kDa
|
|
chr11_-_27637771
|
0.083
|
NM_001143816
NM_170735
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr7_-_38279757
|
0.082
|
NM_001003799
NM_001003806
|
TARP
TRGV9
|
TCR gamma alternate reading frame protein
T cell receptor gamma variable 9
|
|
chr5_+_147423727
|
0.081
|
NM_001127698
NM_001127699
NM_006846
|
SPINK5
|
serine peptidase inhibitor, Kazal type 5
|
|
chr18_+_8707368
|
0.080
|
NM_015210
|
KIAA0802
|
KIAA0802
|
|
chr4_-_69116839
|
0.080
|
NM_001077
|
UGT2B17
|
UDP glucuronosyltransferase 2 family, polypeptide B17
|
|
chr3_+_46894239
|
0.079
|
NM_000316
|
PTH1R
|
parathyroid hormone 1 receptor
|
|
chrX_+_46938178
|
0.079
|
|
UBA1
|
ubiquitin-like modifier activating enzyme 1
|
|
chr3_+_161040439
|
0.079
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr2_+_234191029
|
0.078
|
NM_019076
|
UGT1A8
|
UDP glucuronosyltransferase 1 family, polypeptide A8
|
|
chr16_+_1668257
|
0.078
|
NM_144570
|
HN1L
|
hematological and neurological expressed 1-like
|
|
chr2_+_166137091
|
0.077
|
NM_024969
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr3_+_161040343
|
0.076
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr3_-_121065112
|
0.076
|
|
|
|
|
chrX_+_21960841
|
0.076
|
NM_000444
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked
|
|
chrM_+_4465
|
0.076
|
|
|
|
|
chr3_-_152403636
|
0.075
|
NM_013308
|
GPR171
|
G protein-coupled receptor 171
|
|
chr5_+_140235114
|
0.074
|
NM_018903
NM_031864
|
PCDHA12
|
protocadherin alpha 12
|
|
chr5_+_140552087
|
0.073
|
NM_018930
|
PCDHB10
|
protocadherin beta 10
|
|
chr19_-_21742162
|
0.072
|
NM_173531
|
ZNF100
|
zinc finger protein 100
|