|
chr11_+_133444175
|
3.589
|
|
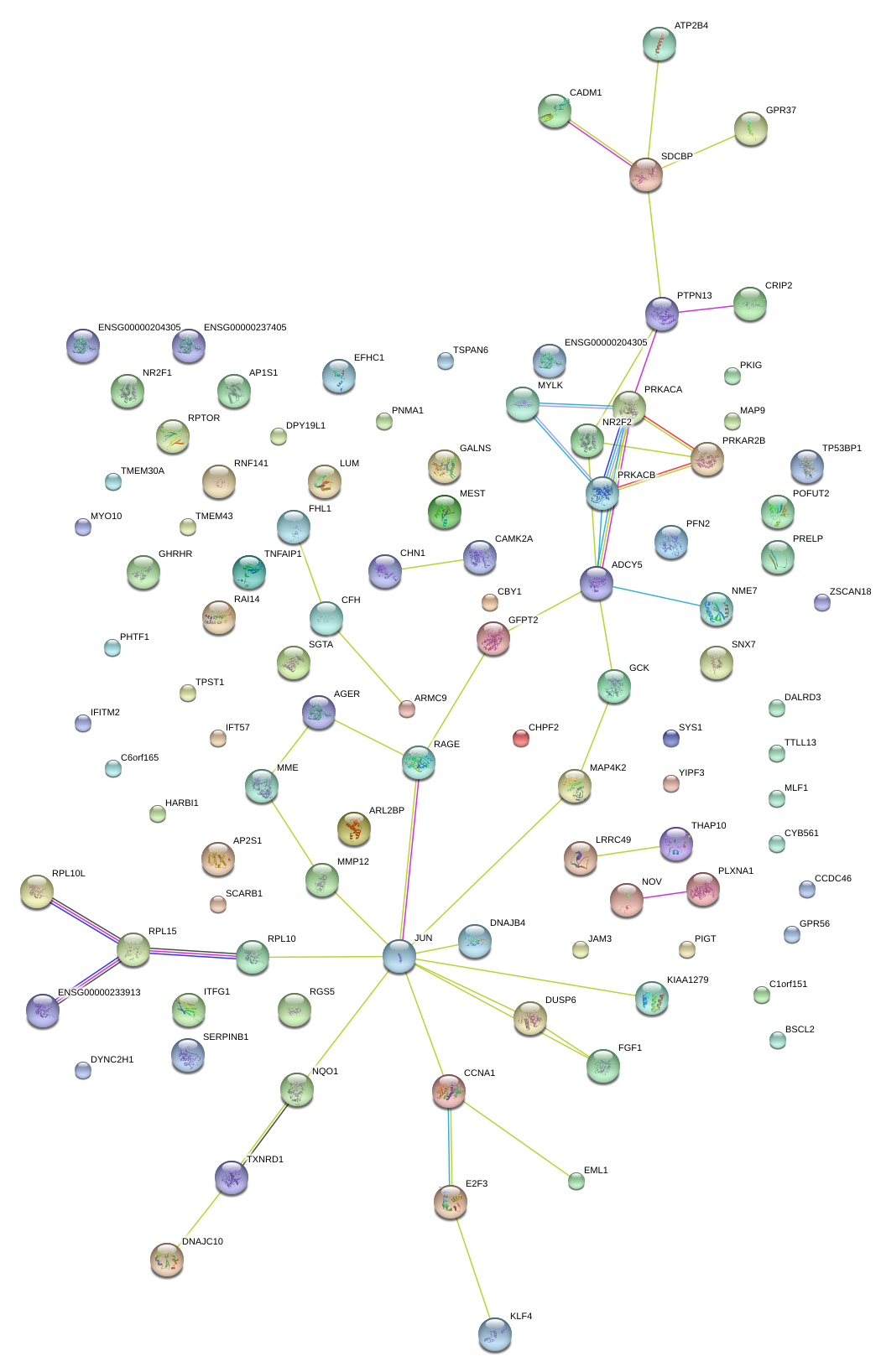
JAM3
|
junctional adhesion molecule 3
|
|
chr11_+_133444029
|
3.392
|
NM_032801
|
JAM3
|
junctional adhesion molecule 3
|
|
chr2_-_175578156
|
2.605
|
NM_001025201
NM_001822
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr15_+_68971835
|
2.365
|
NM_001199017
NM_017691
NM_001199018
|
LRRC49
|
leucine rich repeat containing 49
|
|
chrX_+_135079461
|
2.215
|
NM_001167819
|
FHL1
|
four and a half LIM domains 1
|
|
chrX_+_135079120
|
2.151
|
NM_001159704
|
FHL1
|
four and a half LIM domains 1
|
|
chr15_-_68971665
|
1.939
|
NM_020147
|
THAP10
|
THAP domain containing 10
|
|
chr14_+_105012106
|
1.888
|
NM_001312
|
CRIP2
|
cysteine-rich protein 2
|
|
chr3_-_124822113
|
1.853
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr14_-_73250094
|
1.711
|
NM_006029
|
PNMA1
|
paraneoplastic antigen MA1
|
|
chr4_-_156517423
|
1.702
|
|
MAP9
|
microtubule-associated protein 9
|
|
chr4_-_156517394
|
1.497
|
|
MAP9
|
microtubule-associated protein 9
|
|
chr5_+_34720368
|
1.422
|
NM_001145521
NM_001145525
|
RAI14
|
retinoic acid induced 14
|
|
chr7_+_129919157
|
1.407
|
NM_002402
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr15_+_94674849
|
1.374
|
NM_021005
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr1_+_201862552
|
1.355
|
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4
|
|
chr14_+_99329198
|
1.342
|
NM_001008707
NM_004434
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr1_+_201862548
|
1.338
|
NM_001001396
NM_001684
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4
|
|
chr12_-_88270375
|
1.325
|
NM_001946
NM_022652
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr3_-_124821868
|
1.310
|
|
MYLK
|
myosin light chain kinase
|
|
chr19_-_63301418
|
1.303
|
NM_001145543
NM_001145544
NM_023926
|
ZSCAN18
|
zinc finger and SCAN domain containing 18
|
|
chr5_-_179712920
|
1.302
|
NM_005110
|
GFPT2
|
glutamine-fructose-6-phosphate transaminase 2
|
|
chr16_-_68317855
|
1.269
|
|
NQO1
|
NAD(P)H dehydrogenase, quinone 1
|
|
chrX_-_99778340
|
1.269
|
NM_003270
|
TSPAN6
|
tetraspanin 6
|
|
chr4_-_156517544
|
1.259
|
NM_001039580
|
MAP9
|
microtubule-associated protein 9
|
|
chr3_+_156280129
|
1.240
|
NM_000902
|
MME
|
membrane metallo-endopeptidase
|
|
chr1_-_167603623
|
1.239
|
NM_013330
NM_197972
|
NME7
|
non-metastatic cells 7, protein expressed in (nucleoside-diphosphate kinase)
|
|
chr11_+_102485293
|
1.215
|
NM_001080463
NM_001377
|
DYNC2H1
|
dynein, cytoplasmic 2, heavy chain 1
|
|
chr20_+_43425153
|
1.177
|
NM_001099791
NM_033542
|
SYS1-DBNDD2
SYS1
|
SYS1-DBNDD2 read-through transcript
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae)
|
|
chr16_+_55836480
|
1.139
|
NM_012106
|
ARL2BP
|
ADP-ribosylation factor-like 2 binding protein
|
|
chr16_-_68317868
|
1.112
|
|
NQO1
|
NAD(P)H dehydrogenase, quinone 1
|
|
chr17_-_5344898
|
1.107
|
NM_001162371
|
LOC728392
|
hypothetical protein LOC728392
|
|
chr9_-_109291866
|
1.097
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr1_+_84402962
|
1.095
|
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr12_-_88270128
|
1.070
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr17_-_5345167
|
1.056
|
|
LOC728392
|
hypothetical protein LOC728392
|
|
chr5_-_16795336
|
1.043
|
|
MYO10
|
myosin X
|
|
chr20_+_42593849
|
1.024
|
NM_007066
NM_181804
NM_181805
|
PKIG
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma
|
|
chr1_-_59022275
|
1.019
|
NM_002228
|
JUN
|
jun proto-oncogene
|
|
chr22_+_37382574
|
1.018
|
NM_001002880
NM_015373
|
CBY1
|
chibby homolog 1 (Drosophila)
|
|
chr6_+_52392907
|
1.009
|
NM_018100
|
EFHC1
|
EF-hand domain (C-terminal) containing 1
|
|
chr20_+_43478120
|
0.990
|
NM_001184728
NM_001184729
NM_001184730
NM_015937
|
PIGT
|
phosphatidylinositol glycan anchor biosynthesis, class T
|
|
chr17_-_61618276
|
0.989
|
NM_145036
|
CCDC46
|
coiled-coil domain containing 46
|
|
chr17_+_23686674
|
0.984
|
NM_021137
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial)
|
|
chr4_+_87734489
|
0.980
|
NM_006264
NM_080683
NM_080684
NM_080685
|
PTPN13
|
protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase)
|
|
chr22_+_37382617
|
0.979
|
|
CBY1
|
chibby homolog 1 (Drosophila)
|
|
chr3_-_109423858
|
0.978
|
NM_018010
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas)
|
|
chr7_-_35044177
|
0.971
|
NM_015283
|
DPY19L1
|
dpy-19-like 1 (C. elegans)
|
|
chr8_+_59628372
|
0.968
|
|
SDCBP
|
syndecan binding protein (syntenin)
|
|
chr8_+_59628281
|
0.959
|
NM_001007067
NM_001007068
NM_001007069
NM_001007070
NM_005625
|
SDCBP
|
syndecan binding protein (syntenin)
|
|
chr7_+_150560459
|
0.955
|
NM_019015
|
CHPF2
|
chondroitin polymerizing factor 2
|
|
chr8_+_59628352
|
0.955
|
|
SDCBP
|
syndecan binding protein (syntenin)
|
|
chr3_-_151171576
|
0.931
|
|
PFN2
|
profilin 2
|
|
chr7_+_65307816
|
0.928
|
|
TPST1
|
tyrosylprotein sulfotransferase 1
|
|
chr1_-_114103276
|
0.916
|
NM_006608
|
PHTF1
|
putative homeodomain transcription factor 1
|
|
chr6_+_88174408
|
0.911
|
NM_001031743
|
C6orf165
|
chromosome 6 open reading frame 165
|
|
chr12_-_88270369
|
0.900
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr7_+_65307691
|
0.867
|
NM_003596
|
TPST1
|
tyrosylprotein sulfotransferase 1
|
|
chr1_+_201862845
|
0.855
|
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4
|
|
chr10_+_70418482
|
0.855
|
NM_015634
|
KIAA1279
|
KIAA1279
|
|
chr16_+_56230707
|
0.840
|
NM_001145774
|
GPR56
|
G protein-coupled receptor 56
|
|
chr1_-_114103616
|
0.829
|
|
PHTF1
|
putative homeodomain transcription factor 1
|
|
chr14_-_101841222
|
0.810
|
NM_014226
|
RAGE
|
renal tumor antigen
|
|
chr9_-_109291575
|
0.761
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr1_-_161380562
|
0.749
|
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr12_+_103204842
|
0.744
|
NM_003330
NM_182729
NM_182742
NM_182743
|
TXNRD1
|
thioredoxin reductase 1
|
|
chr12_-_90029672
|
0.734
|
NM_002345
|
LUM
|
lumican
|
|
chr5_-_149649593
|
0.734
|
NM_015981
NM_171825
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr1_+_19796044
|
0.729
|
NM_001032363
|
C1orf15-NBL1
C1orf151
|
C1ORF15-NBL1 read-through transcript
chromosome 1 open reading frame 151
|
|
chr6_-_43592670
|
0.727
|
|
YIPF3
|
Yip1 domain family, member 3
|
|
chr7_+_100584397
|
0.703
|
NM_001283
|
AP1S1
|
adaptor-related protein complex 1, sigma 1 subunit
|
|
chr8_+_120497732
|
0.703
|
NM_002514
|
NOV
|
nephroblastoma overexpressed gene
|
|
chr1_+_98899734
|
0.692
|
NM_015976
NM_152238
|
SNX7
|
sorting nexin 7
|
|
chr2_+_231771623
|
0.690
|
|
ARMC9
|
armadillo repeat containing 9
|
|
chr7_-_44195405
|
0.682
|
NM_000162
|
GCK
|
glucokinase (hexokinase 4)
|
|
chr6_-_76051351
|
0.681
|
NM_001143958
NM_018247
|
TMEM30A
|
transmembrane protein 30A
|
|
chr19_-_52045890
|
0.679
|
NM_004069
NM_021575
|
AP2S1
|
adaptor-related protein complex 2, sigma 1 subunit
|
|
chr3_+_159771646
|
0.675
|
NM_001130156
NM_001130157
NM_001195432
NM_001195433
NM_001195434
NM_022443
|
MLF1
|
myeloid leukemia factor 1
|
|
chr16_-_68317945
|
0.658
|
NM_000903
NM_001025433
NM_001025434
|
NQO1
|
NAD(P)H dehydrogenase, quinone 1
|
|
chr17_-_58877276
|
0.658
|
NM_001017916
NM_001915
|
CYB561
|
cytochrome b-561
|
|
chr17_+_76133337
|
0.658
|
|
RPTOR
|
regulatory associated protein of MTOR, complex 1
|
|
chr12_+_103205148
|
0.643
|
|
TXNRD1
|
thioredoxin reductase 1
|
|
chr6_+_20510019
|
0.638
|
NM_001949
|
E2F3
|
E2F transcription factor 3
|
|
chr15_-_41589958
|
0.629
|
NM_005657
|
TP53BP1
|
tumor protein p53 binding protein 1
|
|
chr11_-_62230078
|
0.616
|
NM_032667
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr1_+_201711578
|
0.607
|
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein
|
|
chr2_+_231771622
|
0.607
|
|
ARMC9
|
armadillo repeat containing 9
|
|
chr16_-_46052254
|
0.603
|
|
ITFG1
|
integrin alpha FG-GAP repeat containing 1
|
|
chr16_-_87450734
|
0.602
|
|
GALNS
|
galactosamine (N-acetyl)-6-sulfate sulfatase
|
|
chr3_+_14141440
|
0.592
|
NM_024334
|
TMEM43
|
transmembrane protein 43
|
|
chr3_-_49030920
|
0.589
|
NM_001009996
|
DALRD3
|
DALR anticodon binding domain containing 3
|
|
chr1_+_78217510
|
0.586
|
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4
|
|
chr2_+_231771584
|
0.583
|
NM_025139
|
ARMC9
|
armadillo repeat containing 9
|
|
chr5_-_142045770
|
0.581
|
NM_000800
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr2_+_183289205
|
0.579
|
NM_018981
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10
|
|
chr15_-_41572558
|
0.578
|
NM_001141979
NM_001141980
|
TP53BP1
|
tumor protein p53 binding protein 1
|
|
chr11_-_10519265
|
0.576
|
NM_016422
|
RNF141
|
ring finger protein 141
|
|
chr21_-_45532176
|
0.576
|
NM_015227
NM_133635
|
POFUT2
|
protein O-fucosyltransferase 2
|
|
chr6_-_2787111
|
0.575
|
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1
|
|
chr13_+_35904408
|
0.575
|
NM_001111045
NM_003914
|
CCNA1
|
cyclin A1
|
|
chr6_-_76051216
|
0.574
|
|
TMEM30A
|
transmembrane protein 30A
|
|
chr7_+_106472243
|
0.571
|
NM_002736
|
PRKAR2B
|
protein kinase, cAMP-dependent, regulatory, type II, beta
|
|
chr15_+_88593765
|
0.568
|
NM_001029964
|
TTLL13
|
tubulin tyrosine ligase-like family, member 13
|
|
chr7_-_124192265
|
0.567
|
|
GPR37
|
G protein-coupled receptor 37 (endothelin receptor type B-like)
|
|
chr5_+_92944680
|
0.563
|
NM_005654
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr6_-_76051197
|
0.553
|
|
TMEM30A
|
transmembrane protein 30A
|
|
chr16_-_46052482
|
0.546
|
NM_030790
|
ITFG1
|
integrin alpha FG-GAP repeat containing 1
|
|
chr11_-_114880275
|
0.545
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chr19_-_2734313
|
0.540
|
|
SGTA
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha
|
|
chr13_-_95092283
|
0.538
|
|
DZIP1
|
DAZ interacting protein 1
|
|
chr13_+_23361591
|
0.531
|
NM_001135816
|
PCOTH
|
prostate collagen triple helix
|
|
chr1_+_167603817
|
0.527
|
NM_003666
|
BLZF1
|
basic leucine zipper nuclear factor 1
|
|
chr19_-_2734332
|
0.524
|
NM_003021
|
SGTA
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha
|
|
chr5_-_149649380
|
0.521
|
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr9_+_115396020
|
0.519
|
NM_144489
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr1_-_181871490
|
0.519
|
|
ARPC5
|
actin related protein 2/3 complex, subunit 5, 16kDa
|
|
chr1_+_37795101
|
0.519
|
NM_003462
|
DNALI1
|
dynein, axonemal, light intermediate chain 1
|
|
chr8_-_102033409
|
0.516
|
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr8_-_98359347
|
0.511
|
NM_033512
|
TSPYL5
|
TSPY-like 5
|
|
chr16_-_56393871
|
0.510
|
|
KIFC3
|
kinesin family member C3
|
|
chr11_+_131285747
|
0.506
|
NM_001144058
NM_001144059
NM_016522
|
NTM
|
neurotrimin
|
|
chr7_-_139987044
|
0.505
|
|
DENND2A
|
DENN/MADD domain containing 2A
|
|
chr4_+_153920563
|
0.505
|
|
ARFIP1
|
ADP-ribosylation factor interacting protein 1
|
|
chr13_-_35818491
|
0.504
|
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr5_-_142046175
|
0.502
|
NM_033136
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr6_-_2787068
|
0.501
|
NM_030666
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1
|
|
chr4_+_153920382
|
0.497
|
NM_001025593
NM_001025595
NM_014447
|
ARFIP1
|
ADP-ribosylation factor interacting protein 1
|
|
chr7_+_138469029
|
0.496
|
NM_001144920
NM_001144923
NM_024926
|
TTC26
|
tetratricopeptide repeat domain 26
|
|
chr11_-_33752468
|
0.487
|
NM_012175
NM_033406
|
FBXO3
|
F-box protein 3
|
|
chr3_-_10337837
|
0.480
|
NM_183352
|
SEC13
|
SEC13 homolog (S. cerevisiae)
|
|
chr15_+_99237555
|
0.479
|
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3
|
|
chr3_-_49736297
|
0.475
|
NM_013334
NM_021971
|
GMPPB
|
GDP-mannose pyrophosphorylase B
|
|
chr12_-_129889569
|
0.473
|
NM_001980
NM_194356
|
STX2
|
syntaxin 2
|
|
chr8_-_98359248
|
0.468
|
|
TSPYL5
|
TSPY-like 5
|
|
chr1_-_181871416
|
0.467
|
|
ARPC5
|
actin related protein 2/3 complex, subunit 5, 16kDa
|
|
chr9_+_131855805
|
0.464
|
NM_001136557
NM_001136558
NM_020960
|
GPR107
|
G protein-coupled receptor 107
|
|
chr22_-_41815277
|
0.463
|
NM_012263
|
TTLL1
|
tubulin tyrosine ligase-like family, member 1
|
|
chr19_+_43971689
|
0.459
|
NM_001042507
|
LGALS7B
|
lectin, galactoside-binding, soluble, 7B
|
|
chr10_+_99390431
|
0.457
|
NM_018425
|
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha
|
|
chr2_-_9688552
|
0.456
|
NM_006826
|
YWHAQ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide
|
|
chr3_-_10337724
|
0.448
|
NM_001136232
|
SEC13
|
SEC13 homolog (S. cerevisiae)
|
|
chr19_-_2734225
|
0.448
|
|
SGTA
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha
|
|
chr1_-_44593510
|
0.444
|
|
ERI3
|
ERI1 exoribonuclease family member 3
|
|
chr7_-_22506320
|
0.440
|
NM_001164460
NM_207342
|
MGC87042
|
STEAP family protein MGC87042
|
|
chr11_-_65869113
|
0.439
|
NM_001024957
NM_015399
|
BRMS1
|
breast cancer metastasis suppressor 1
|
|
chr16_-_56393881
|
0.438
|
NM_001130100
NM_005550
|
KIFC3
|
kinesin family member C3
|
|
chrX_+_48341496
|
0.436
|
|
WDR13
|
WD repeat domain 13
|
|
chr6_-_43592679
|
0.435
|
NM_015388
|
YIPF3
|
Yip1 domain family, member 3
|
|
chrX_+_48341164
|
0.434
|
NM_001166426
|
WDR13
|
WD repeat domain 13
|
|
chr10_-_17283623
|
0.433
|
|
TRDMT1
|
tRNA aspartic acid methyltransferase 1
|
|
chr3_-_10337776
|
0.429
|
|
SEC13
|
SEC13 homolog (S. cerevisiae)
|
|
chr19_-_55062611
|
0.429
|
NM_007254
|
PNKP
|
polynucleotide kinase 3'-phosphatase
|
|
chr9_-_34610489
|
0.426
|
NM_007234
NM_024348
|
DCTN3
|
dynactin 3 (p22)
|
|
chr8_+_22518218
|
0.426
|
|
KIAA1967
|
KIAA1967
|
|
chr1_-_44593287
|
0.424
|
|
ERI3
|
ERI1 exoribonuclease family member 3
|
|
chrX_+_129301493
|
0.423
|
NM_003951
NM_022810
|
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14
|
|
chr18_+_52469559
|
0.421
|
NM_015285
NM_052834
|
WDR7
|
WD repeat domain 7
|
|
chr1_-_167663278
|
0.421
|
NM_021179
|
C1orf114
|
chromosome 1 open reading frame 114
|
|
chr19_-_51666629
|
0.417
|
NM_001103149
NM_018215
|
PNMAL1
|
PNMA-like 1
|
|
chr6_-_43592481
|
0.417
|
|
YIPF3
|
Yip1 domain family, member 3
|
|
chr1_-_44593522
|
0.416
|
NM_024066
|
ERI3
|
ERI1 exoribonuclease family member 3
|
|
chr9_-_34610483
|
0.416
|
|
DCTN3
|
dynactin 3 (p22)
|
|
chr15_-_82998516
|
0.413
|
NM_032856
|
WDR73
|
WD repeat domain 73
|
|
chr15_-_53487748
|
0.411
|
NM_004748
NM_020739
|
CCPG1
|
cell cycle progression 1
|
|
chr8_-_102033444
|
0.409
|
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr14_+_73555802
|
0.404
|
NM_025057
|
C14orf45
|
chromosome 14 open reading frame 45
|
|
chr14_-_22127902
|
0.403
|
|
DAD1
|
defender against cell death 1
|
|
chr1_+_207926172
|
0.402
|
NM_181755
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1
|
|
chr14_-_22127954
|
0.402
|
NM_001344
|
DAD1
|
defender against cell death 1
|
|
chr9_-_34610460
|
0.398
|
|
DCTN3
|
dynactin 3 (p22)
|
|
chr15_-_82998500
|
0.397
|
|
WDR73
|
WD repeat domain 73
|
|
chr8_+_22518231
|
0.394
|
|
KIAA1967
|
KIAA1967
|
|
chr21_-_33935883
|
0.392
|
NM_145858
|
CRYZL1
|
crystallin, zeta (quinone reductase)-like 1
|
|
chr8_+_22518212
|
0.387
|
|
KIAA1967
|
KIAA1967
|
|
chr6_-_32271868
|
0.385
|
|
NOTCH4
|
notch 4
|
|
chr2_-_9688450
|
0.385
|
|
YWHAQ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide
|
|
chrX_+_48341066
|
0.384
|
|
WDR13
|
WD repeat domain 13
|
|
chr8_+_9450849
|
0.384
|
NM_003747
|
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase
|
|
chr16_+_56219802
|
0.383
|
NM_001145771
NM_001145772
NM_001145773
NM_201524
NM_201525
|
GPR56
|
G protein-coupled receptor 56
|
|
chr1_-_181871473
|
0.381
|
|
ARPC5
|
actin related protein 2/3 complex, subunit 5, 16kDa
|
|
chr19_+_3523958
|
0.381
|
|
HMG20B
|
high-mobility group 20B
|
|
chr1_+_109458093
|
0.379
|
NM_020775
|
KIAA1324
|
KIAA1324
|
|
chr5_+_148501178
|
0.378
|
NM_014945
|
ABLIM3
|
actin binding LIM protein family, member 3
|
|
chr2_+_170044194
|
0.378
|
NM_152384
|
BBS5
|
Bardet-Biedl syndrome 5
|
|
chr14_-_23734598
|
0.377
|
NM_001014842
NM_006405
|
TM9SF1
|
transmembrane 9 superfamily member 1
|
|
chr7_-_102707901
|
0.377
|
|
DPY19L2P2
|
dpy-19-like 2 pseudogene 2 (C. elegans)
|
|
chr3_+_62279707
|
0.377
|
|
C3orf14
|
chromosome 3 open reading frame 14
|
|
chr1_+_201711505
|
0.375
|
NM_002725
NM_201348
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein
|
|
chr16_-_46052596
|
0.373
|
|
ITFG1
|
integrin alpha FG-GAP repeat containing 1
|
|
chr8_+_22518477
|
0.372
|
NM_021174
|
KIAA1967
|
KIAA1967
|
|
chr2_-_47257156
|
0.371
|
|
CALM2
|
calmodulin 2 (phosphorylase kinase, delta)
|
|
chr14_-_23734433
|
0.371
|
|
TM9SF1
|
transmembrane 9 superfamily member 1
|
|
chr3_-_58538110
|
0.370
|
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chr3_+_132228383
|
0.370
|
NM_001146003
NM_024800
NM_145910
|
NEK11
|
NIMA (never in mitosis gene a)- related kinase 11
|
|
chr2_-_47257183
|
0.368
|
NM_001743
|
CALM2
|
calmodulin 2 (phosphorylase kinase, delta)
|
|
chr19_+_44308224
|
0.368
|
NM_001014831
NM_001014832
NM_001014834
NM_001014835
NM_005884
|
PAK4
|
p21 protein (Cdc42/Rac)-activated kinase 4
|
|
chr14_+_101899104
|
0.366
|
|
TECPR2
|
tectonin beta-propeller repeat containing 2
|
|
chr6_-_158985727
|
0.366
|
NM_006519
|
DYNLT1
|
dynein, light chain, Tctex-type 1
|
|
chr2_-_47257150
|
0.365
|
|
CALM2
|
calmodulin 2 (phosphorylase kinase, delta)
|
|
chr1_-_44593526
|
0.365
|
|
ERI3
|
ERI1 exoribonuclease family member 3
|