|
chrX_-_151688850
|
3.420
|
NM_005362
|
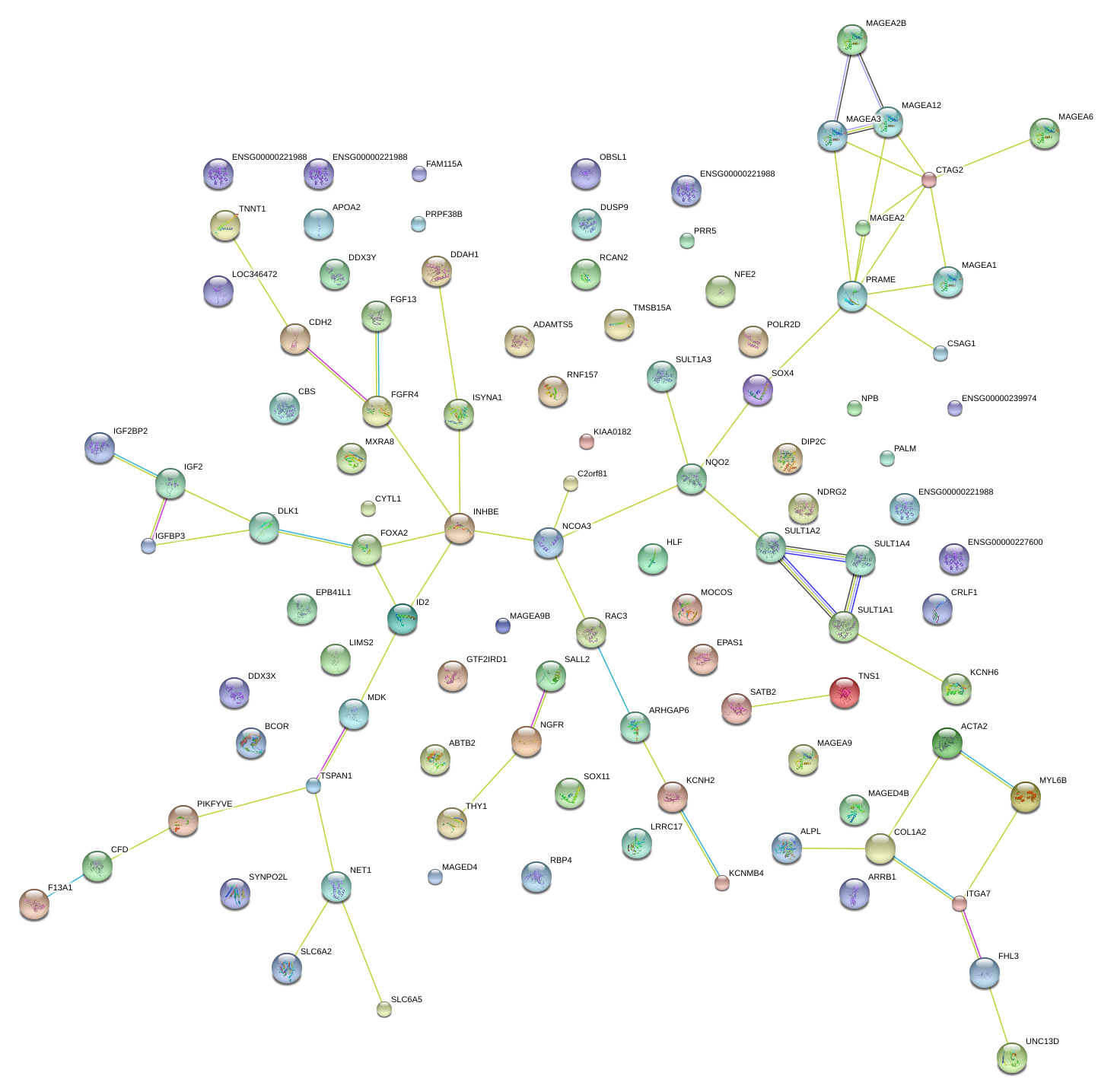
MAGEA3
|
melanoma antigen family A, 3
|
|
chrX_+_151617900
|
3.352
|
NM_005363
NM_175868
|
MAGEA6
|
melanoma antigen family A, 6
|
|
chr11_+_46359855
|
2.985
|
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chrX_-_151653756
|
2.913
|
NM_001166386
NM_001166387
NM_005367
|
MAGEA12
|
melanoma antigen family A, 12
|
|
chr11_+_46359821
|
2.877
|
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr11_+_46359836
|
2.754
|
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chrX_-_152139267
|
2.447
|
NM_004988
|
MAGEA1
|
melanoma antigen family A, 1 (directs expression of antigen MZ2-E)
|
|
chr11_+_46359781
|
2.368
|
NM_002391
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr11_+_46359878
|
2.216
|
NM_001012333
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr19_-_60350122
|
2.197
|
|
TNNT1
|
troponin T type 1 (skeletal, slow)
|
|
chr2_-_218551786
|
2.163
|
|
TNS1
|
tensin 1
|
|
chrX_+_151653874
|
1.910
|
NM_001102576
NM_153478
|
CSAG1
|
chondrosarcoma associated gene 1
|
|
chr20_-_22512893
|
1.720
|
|
FOXA2
|
forkhead box A2
|
|
chrX_+_151633730
|
1.697
|
NM_005361
NM_175742
NM_175743
|
MAGEA2
|
melanoma antigen family A, 2
|
|
chr21_-_27261276
|
1.674
|
NM_007038
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5
|
|
chrX_+_152561086
|
1.611
|
NM_001395
|
DUSP9
|
dual specificity phosphatase 9
|
|
chrX_-_137621492
|
1.607
|
NM_004114
|
FGF13
|
fibroblast growth factor 13
|
|
chr19_-_18578577
|
1.567
|
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr19_-_18578631
|
1.560
|
NM_004750
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr6_+_2945025
|
1.555
|
NM_000904
|
NQO2
|
NAD(P)H dehydrogenase, quinone 2
|
|
chr7_-_45927263
|
1.538
|
NM_000598
NM_001013398
|
IGFBP3
|
insulin-like growth factor binding protein 3
|
|
chrX_-_151672985
|
1.497
|
NM_005361
NM_175742
NM_175743
|
MAGEA2
|
melanoma antigen family A, 2
|
|
chr19_-_60350118
|
1.463
|
|
TNNT1
|
troponin T type 1 (skeletal, slow)
|
|
chr19_-_18578467
|
1.443
|
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr19_-_60352369
|
1.440
|
NM_001126132
NM_001126133
NM_003283
|
TNNT1
|
troponin T type 1 (skeletal, slow)
|
|
chr16_+_84202515
|
1.409
|
NM_001134473
|
KIAA0182
|
KIAA0182
|
|
chrX_-_137621060
|
1.371
|
|
FGF13
|
fibroblast growth factor 13
|
|
chr21_-_43368963
|
1.356
|
NM_000071
|
CBS
|
cystathionine-beta-synthase
|
|
chr21_-_43369492
|
1.318
|
NM_001178008
NM_001178009
|
CBS
|
cystathionine-beta-synthase
|
|
chr1_-_159459814
|
1.214
|
|
APOA2
|
apolipoprotein A-II
|
|
chr11_-_2108035
|
1.197
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chrX_-_39841599
|
1.193
|
NM_001123385
NM_017745
|
BCOR
|
BCL6 corepressor
|
|
chr19_+_811615
|
1.178
|
|
CFD
|
complement factor D (adipsin)
|
|
chr3_-_187025501
|
1.157
|
NM_001007225
NM_006548
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr7_+_93861808
|
1.148
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr11_-_74740298
|
1.130
|
NM_004041
NM_020251
|
ARRB1
|
arrestin, beta 1
|
|
chr6_-_6265896
|
1.127
|
NM_000129
|
F13A1
|
coagulation factor XIII, A1 polypeptide
|
|
chr2_-_128149112
|
1.116
|
NM_001136037
NM_001161403
|
LIMS2
|
LIM and senescent cell antigen-like domains 2
|
|
chr22_-_21231421
|
1.103
|
NM_206955
NM_206956
NM_006115
NM_206953
NM_206954
|
PRAME
|
preferentially expressed antigen in melanoma
|
|
chr11_-_2115086
|
1.096
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr1_-_1283770
|
1.091
|
NM_032348
|
MXRA8
|
matrix-remodelling associated 8
|
|
chr20_-_22513050
|
1.059
|
NM_021784
|
FOXA2
|
forkhead box A2
|
|
chr2_+_8739563
|
1.014
|
NM_002166
|
ID2
|
inhibitor of DNA binding 2, dominant negative helix-loop-helix protein
|
|
chr7_-_150305933
|
0.995
|
NM_000238
NM_172056
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr10_-_75085754
|
0.982
|
NM_001114133
|
SYNPO2L
|
synaptopodin 2-like
|
|
chr6_-_6265874
|
0.968
|
|
F13A1
|
coagulation factor XIII, A1 polypeptide
|
|
chr16_-_28515868
|
0.963
|
NM_001054
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2
|
|
chr5_+_176446425
|
0.940
|
NM_002011
NM_213647
|
FGFR4
|
fibroblast growth factor receptor 4
|
|
chr12_+_69046241
|
0.934
|
NM_014505
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4
|
|
chrX_+_51944658
|
0.931
|
NM_001098800
NM_030801
NM_177535
NM_177537
|
MAGED4
MAGED4B
|
melanoma antigen family D, 4
melanoma antigen family D, 4B
|
|
chr12_-_54387732
|
0.915
|
|
ITGA7
|
integrin, alpha 7
|
|
chr16_-_28528780
|
0.895
|
NM_001055
NM_177529
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1
|
|
chr1_+_21708428
|
0.894
|
NM_000478
NM_001127501
NM_001177520
|
ALPL
|
alkaline phosphatase, liver/bone/kidney
|
|
chrY_+_13526169
|
0.891
|
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr3_-_187025438
|
0.886
|
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr2_-_220144511
|
0.875
|
NM_001173408
NM_001173431
NM_015311
|
OBSL1
|
obscurin-like 1
|
|
chr16_+_84204424
|
0.873
|
NM_014615
|
KIAA0182
|
KIAA0182
|
|
chrX_-_151037066
|
0.854
|
NM_021049
|
MAGEA5
|
melanoma antigen family A, 5
|
|
chr6_+_32229771
|
0.851
|
|
PPT2-EGFL8
PPT2
|
PPT2-EGFL8 read-through transcript
palmitoyl-protein thioesterase 2
|
|
chr16_+_30118036
|
0.843
|
NM_001017390
NM_177552
|
SULT1A4
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3
|
|
chrX_-_101658297
|
0.835
|
NM_021992
|
TMSB15A
TMSB15B
|
thymosin beta 15a
thymosin beta 15B
|
|
chr19_-_18409788
|
0.828
|
NM_001170939
|
ISYNA1
|
inositol-3-phosphate synthase 1
|
|
chr18_-_24011349
|
0.814
|
NM_001792
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr12_-_52980984
|
0.809
|
NM_001136023
|
NFE2
|
nuclear factor (erythroid-derived 2), 45kDa
|
|
chr12_-_52980914
|
0.809
|
|
NFE2
|
nuclear factor (erythroid-derived 2), 45kDa
|
|
chr7_-_143230096
|
0.798
|
NM_014719
|
FAM115A
|
family with sequence similarity 115, member A
|
|
chr17_+_16225130
|
0.792
|
|
|
|
|
chr6_+_32230172
|
0.791
|
|
PPT2
|
palmitoyl-protein thioesterase 2
|
|
chr22_+_43442987
|
0.787
|
NM_001017528
NM_001017529
|
PRR5
|
proline rich 5 (renal)
|
|
chr10_+_5444516
|
0.783
|
NM_001047160
|
NET1
|
neuroepithelial cell transforming 1
|
|
chr10_-_95350949
|
0.780
|
NM_006744
|
RBP4
|
retinol binding protein 4, plasma
|
|
chr12_+_54832470
|
0.779
|
NM_002475
|
MYL6B
|
myosin, light chain 6B, alkali, smooth muscle and non-muscle
|
|
chrX_-_148476903
|
0.776
|
NM_005365
NM_001080790
|
MAGEA9
MAGEA9B
|
melanoma antigen family A, 9
melanoma antigen family A, 9B
|
|
chrX_-_153534951
|
0.765
|
NM_020994
NM_172377
|
CTAG2
|
cancer/testis antigen 2
|
|
chrY_+_13526092
|
0.764
|
NM_004660
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr11_-_118799058
|
0.761
|
|
THY1
|
Thy-1 cell surface antigen
|
|
chr18_+_32021418
|
0.744
|
NM_017947
|
MOCOS
|
molybdenum cofactor sulfurase
|
|
chr19_-_18409971
|
0.744
|
NM_001170938
NM_016368
|
ISYNA1
|
inositol-3-phosphate synthase 1
|
|
chr11_-_2109536
|
0.744
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr17_+_50697355
|
0.731
|
|
HLF
|
hepatic leukemia factor
|
|
chr10_-_725522
|
0.730
|
NM_014974
|
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila)
|
|
chr7_+_73506047
|
0.729
|
NM_016328
|
GTF2IRD1
|
GTF2I repeat domain containing 1
|
|
chr14_+_100270222
|
0.723
|
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chr7_+_102340579
|
0.720
|
NM_001031692
NM_005824
|
LRRC17
|
leucine rich repeat containing 17
|
|
chr2_-_220144427
|
0.718
|
|
OBSL1
|
obscurin-like 1
|
|
chr6_-_46566969
|
0.714
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chr19_+_659766
|
0.713
|
NM_001040134
NM_002579
|
PALM
|
paralemmin
|
|
chr14_-_20563698
|
0.709
|
|
NDRG2
|
NDRG family member 2
|
|
chr2_-_200030944
|
0.704
|
NM_001172509
|
SATB2
|
SATB homeobox 2
|
|
chr12_+_56135353
|
0.703
|
NM_031479
|
INHBE
|
inhibin, beta E
|
|
chr7_+_73506172
|
0.698
|
|
GTF2IRD1
|
GTF2I repeat domain containing 1
|
|
chr11_-_34336102
|
0.697
|
NM_145804
|
ABTB2
|
ankyrin repeat and BTB (POZ) domain containing 2
|
|
chr18_-_24010944
|
0.689
|
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr12_-_54387934
|
0.682
|
NM_001144996
NM_002206
|
ITGA7
|
integrin, alpha 7
|
|
chrX_-_11194015
|
0.678
|
NM_013423
|
ARHGAP6
|
Rho GTPase activating protein 6
|
|
chr2_+_5750229
|
0.676
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chrX_+_148671394
|
0.672
|
NM_005365
NM_001080790
|
MAGEA9
MAGEA9B
|
melanoma antigen family A, 9
melanoma antigen family A, 9B
|
|
chr7_-_150306169
|
0.666
|
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr1_-_85816520
|
0.666
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr14_-_21064182
|
0.663
|
|
SALL2
|
sal-like 2 (Drosophila)
|
|
chr20_+_34206070
|
0.655
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr14_-_20563722
|
0.649
|
NM_016250
NM_201535
NM_201536
NM_201537
|
NDRG2
|
NDRG family member 2
|
|
chr17_+_44927591
|
0.647
|
NM_002507
|
NGFR
|
nerve growth factor receptor
|
|
chr1_-_38243716
|
0.647
|
|
FHL3
|
four and a half LIM domains 3
|
|
chr16_+_29378694
|
0.643
|
NM_001017390
NM_177552
|
SULT1A4
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3
|
|
chr17_+_77582774
|
0.642
|
NM_005052
|
RAC3
|
ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3)
|
|
chr7_-_92303812
|
0.639
|
|
CDK6
|
cyclin-dependent kinase 6
|
|
chr19_+_14405216
|
0.637
|
|
PKN1
|
protein kinase N1
|
|
chr5_-_39460687
|
0.637
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr1_-_32574185
|
0.631
|
NM_023009
|
MARCKSL1
|
MARCKS-like 1
|
|
chr17_+_50697319
|
0.628
|
NM_002126
|
HLF
|
hepatic leukemia factor
|
|
chr21_+_46367216
|
0.628
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr17_-_72218535
|
0.616
|
NM_001008528
NM_001008529
NM_198530
|
MXRA7
|
matrix-remodelling associated 7
|
|
chr10_+_114699953
|
0.604
|
NM_001146274
NM_001146283
NM_001146284
NM_001146285
NM_001146286
NM_001198525
NM_001198526
NM_001198527
NM_001198528
NM_001198529
NM_001198530
NM_001198531
NM_030756
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr3_+_8750485
|
0.601
|
NM_033337
NM_001234
|
CAV3
|
caveolin 3
|
|
chr11_-_118799452
|
0.600
|
NM_006288
|
THY1
|
Thy-1 cell surface antigen
|
|
chr1_-_38243747
|
0.599
|
NM_004468
|
FHL3
|
four and a half LIM domains 3
|
|
chrX_-_153500584
|
0.596
|
NM_001327
NM_139250
|
CTAG1B
CTAG1A
|
cancer/testis antigen 1B
cancer/testis antigen 1A
|
|
chr20_-_47238082
|
0.590
|
|
STAU1
|
staufen, RNA binding protein, homolog 1 (Drosophila)
|
|
chr4_-_5941043
|
0.586
|
|
CRMP1
|
collapsin response mediator protein 1
|
|
chrX_+_153466611
|
0.582
|
NM_001327
NM_139250
|
CTAG1B
CTAG1A
|
cancer/testis antigen 1B
cancer/testis antigen 1A
|
|
chr17_-_72218661
|
0.580
|
|
MXRA7
|
matrix-remodelling associated 7
|
|
chr1_-_159459999
|
0.580
|
NM_001643
|
APOA2
|
apolipoprotein A-II
|
|
chr7_-_149669622
|
0.580
|
NM_002889
|
RARRES2
|
retinoic acid receptor responder (tazarotene induced) 2
|
|
chr1_+_212227902
|
0.579
|
|
PROX1
|
prospero homeobox 1
|
|
chr2_-_227372574
|
0.578
|
|
IRS1
|
insulin receptor substrate 1
|
|
chr19_+_876984
|
0.574
|
NM_005224
|
ARID3A
|
AT rich interactive domain 3A (BRIGHT-like)
|
|
chr17_-_7023283
|
0.573
|
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chr5_-_176833197
|
0.573
|
|
DBN1
|
drebrin 1
|
|
chr17_-_77474597
|
0.570
|
NM_032711
|
MAFG
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian)
|
|
chr3_+_51984076
|
0.557
|
NM_015407
|
ABHD14A-ACY1
ABHD14A
|
ABHD14A-ACY1 read-through transcript
abhydrolase domain containing 14A
|
|
chr7_-_92303865
|
0.555
|
NM_001145306
|
CDK6
|
cyclin-dependent kinase 6
|
|
chr10_-_76538859
|
0.554
|
NM_001007271
NM_001007272
NM_001007273
|
DUSP13
|
dual specificity phosphatase 13
|
|
chr1_-_21850874
|
0.548
|
NM_001145658
|
RAP1GAP
|
RAP1 GTPase activating protein
|
|
chr19_-_50977485
|
0.547
|
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr7_-_100077072
|
0.546
|
NM_003227
|
TFR2
|
transferrin receptor 2
|
|
chr11_+_124539768
|
0.544
|
NM_022062
|
PKNOX2
|
PBX/knotted 1 homeobox 2
|
|
chr13_+_112808102
|
0.544
|
NM_000131
NM_019616
|
F7
|
coagulation factor VII (serum prothrombin conversion accelerator)
|
|
chr2_-_119322228
|
0.540
|
NM_001426
|
EN1
|
engrailed homeobox 1
|
|
chr2_-_161058550
|
0.538
|
NM_002897
NM_016836
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr1_-_154214522
|
0.533
|
NM_004723
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2
|
|
chr3_+_139550368
|
0.533
|
|
MRAS
|
muscle RAS oncogene homolog
|
|
chr19_+_50141068
|
0.532
|
NM_000483
|
APOC2
|
apolipoprotein C-II
|
|
chr17_-_44058612
|
0.527
|
NM_024017
|
HOXB9
|
homeobox B9
|
|
chr6_-_35572694
|
0.524
|
NM_003214
|
TEAD3
|
TEA domain family member 3
|
|
chr20_+_30813851
|
0.513
|
NM_006892
NM_175848
NM_175849
|
DNMT3B
|
DNA (cytosine-5-)-methyltransferase 3 beta
|
|
chr1_-_149214063
|
0.509
|
NM_022075
NM_181746
|
LASS2
|
LAG1 homolog, ceramide synthase 2
|
|
chr19_-_50977592
|
0.509
|
NM_001081560
NM_001081562
NM_004409
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr11_+_46358909
|
0.503
|
NM_001012334
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr15_+_61122139
|
0.503
|
|
TPM1
|
tropomyosin 1 (alpha)
|
|
chr10_+_22650145
|
0.502
|
|
BMI1
|
BMI1 polycomb ring finger oncogene
|
|
chr1_-_149213916
|
0.499
|
|
LASS2
|
LAG1 homolog, ceramide synthase 2
|
|
chr16_+_70217564
|
0.499
|
NM_001017967
NM_052858
|
MARVELD3
|
MARVEL domain containing 3
|
|
chr12_+_55808514
|
0.498
|
NM_002332
|
LRP1
|
low density lipoprotein receptor-related protein 1
|
|
chr12_-_6586811
|
0.497
|
NM_001273
|
CHD4
|
chromodomain helicase DNA binding protein 4
|
|
chr11_+_124539814
|
0.496
|
|
PKNOX2
|
PBX/knotted 1 homeobox 2
|
|
chrX_+_152562013
|
0.496
|
|
DUSP9
|
dual specificity phosphatase 9
|
|
chr6_+_33486750
|
0.494
|
NM_002636
NM_024165
|
PHF1
|
PHD finger protein 1
|
|
chr7_-_130891861
|
0.491
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr7_+_65731289
|
0.489
|
NM_001167961
NM_153033
|
KCTD7
|
potassium channel tetramerisation domain containing 7
|
|
chr3_+_12304332
|
0.489
|
NM_005037
NM_138712
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr11_-_8695974
|
0.489
|
|
ST5
|
suppression of tumorigenicity 5
|
|
chr2_-_144994349
|
0.487
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr21_+_42946719
|
0.486
|
NM_001001567
NM_001001568
NM_001001569
NM_001001570
NM_001001571
NM_001001572
NM_001001573
NM_001001574
NM_001001575
NM_001001576
NM_001001577
NM_001001578
NM_001001579
NM_001001580
NM_001001581
NM_001001582
NM_001001583
NM_001001584
NM_001001585
NM_002606
|
PDE9A
|
phosphodiesterase 9A
|
|
chr20_+_30814048
|
0.483
|
|
DNMT3B
|
DNA (cytosine-5-)-methyltransferase 3 beta
|
|
chr12_-_69600514
|
0.479
|
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr9_-_126945536
|
0.479
|
NM_001144877
NM_173690
|
SCAI
|
suppressor of cancer cell invasion
|
|
chr4_-_186693574
|
0.478
|
NM_001114107
NM_014476
|
PDLIM3
|
PDZ and LIM domain 3
|
|
chr1_-_149213929
|
0.478
|
|
LASS2
|
LAG1 homolog, ceramide synthase 2
|
|
chr7_-_19123787
|
0.476
|
NM_000474
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr3_+_156280338
|
0.475
|
NM_007287
|
MME
|
membrane metallo-endopeptidase
|
|
chr6_-_2916980
|
0.474
|
|
SERPINB6
|
serpin peptidase inhibitor, clade B (ovalbumin), member 6
|
|
chr2_-_128149800
|
0.473
|
|
LIMS2
|
LIM and senescent cell antigen-like domains 2
|
|
chrX_-_48578878
|
0.471
|
NM_013271
|
PCSK1N
|
proprotein convertase subtilisin/kexin type 1 inhibitor
|
|
chr10_+_21863099
|
0.468
|
NM_001195627
NM_001195628
NM_001195630
NM_004641
|
MLLT10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10
|
|
chr19_+_14405100
|
0.468
|
NM_002741
|
PKN1
|
protein kinase N1
|
|
chr5_-_111121150
|
0.467
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr7_+_150390566
|
0.464
|
|
SLC4A2
|
solute carrier family 4, anion exchanger, member 2 (erythrocyte membrane protein band 3-like 1)
|
|
chr11_-_404975
|
0.463
|
NM_001135054
|
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain
|
|
chr2_+_46779553
|
0.461
|
NM_014011
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr5_-_77626222
|
0.461
|
|
AP3B1
|
adaptor-related protein complex 3, beta 1 subunit
|
|
chr12_+_26239535
|
0.455
|
NM_001135823
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr17_-_74432800
|
0.453
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr19_+_40321584
|
0.452
|
|
FXYD1
|
FXYD domain containing ion transport regulator 1
|
|
chr19_+_40321567
|
0.452
|
NM_021902
|
FXYD1
|
FXYD domain containing ion transport regulator 1
|
|
chr2_-_158440587
|
0.449
|
NM_001111067
|
ACVR1
|
activin A receptor, type I
|
|
chr17_-_7023378
|
0.447
|
NM_001197216
NM_001671
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chr2_-_158439868
|
0.445
|
NM_001105
|
ACVR1
|
activin A receptor, type I
|
|
chr7_+_73506333
|
0.445
|
NM_005685
|
GTF2IRD1
|
GTF2I repeat domain containing 1
|
|
chr1_-_201422450
|
0.443
|
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chr16_+_166678
|
0.443
|
NM_000558
|
HBA1
HBA2
|
hemoglobin, alpha 1
hemoglobin, alpha 2
|
|
chr21_-_43671355
|
0.442
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr14_-_103058865
|
0.440
|
NM_001823
|
CKB
|
creatine kinase, brain
|
|
chr4_-_5941167
|
0.440
|
NM_001313
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr1_-_149213826
|
0.437
|
|
LASS2
|
LAG1 homolog, ceramide synthase 2
|
|
chr1_-_149213778
|
0.434
|
|
LASS2
|
LAG1 homolog, ceramide synthase 2
|
|
chr7_-_148212312
|
0.433
|
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr1_-_23977330
|
0.433
|
|
LOC100506963
|
hypothetical LOC100506963
|
|
chr15_+_41597097
|
0.430
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr20_+_34143983
|
0.429
|
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|