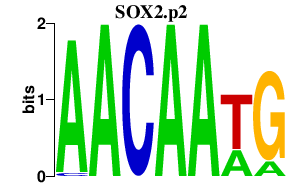
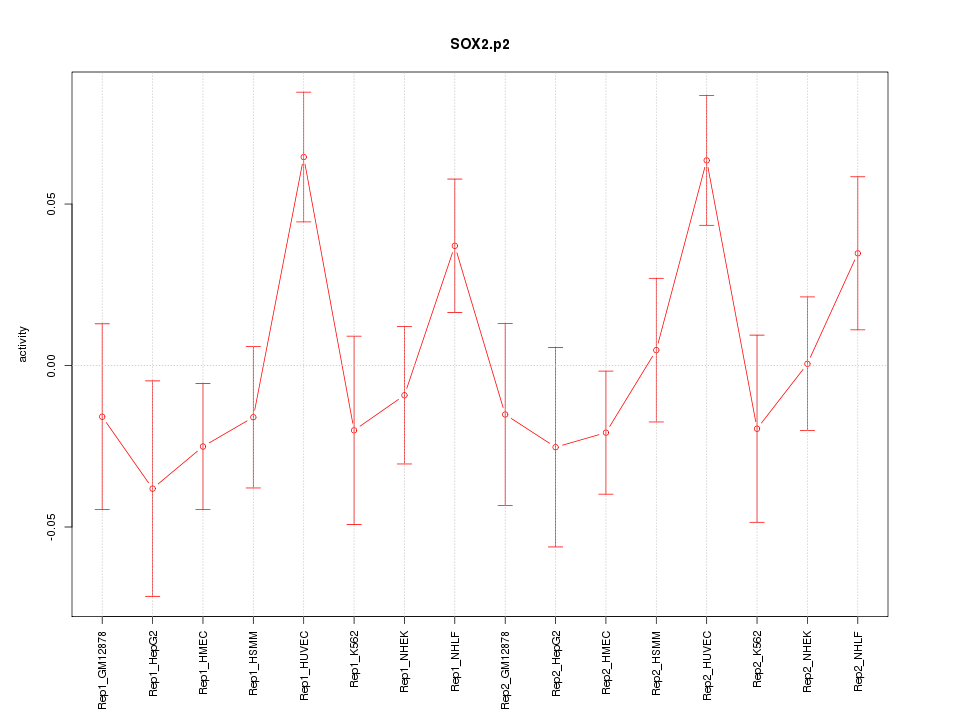
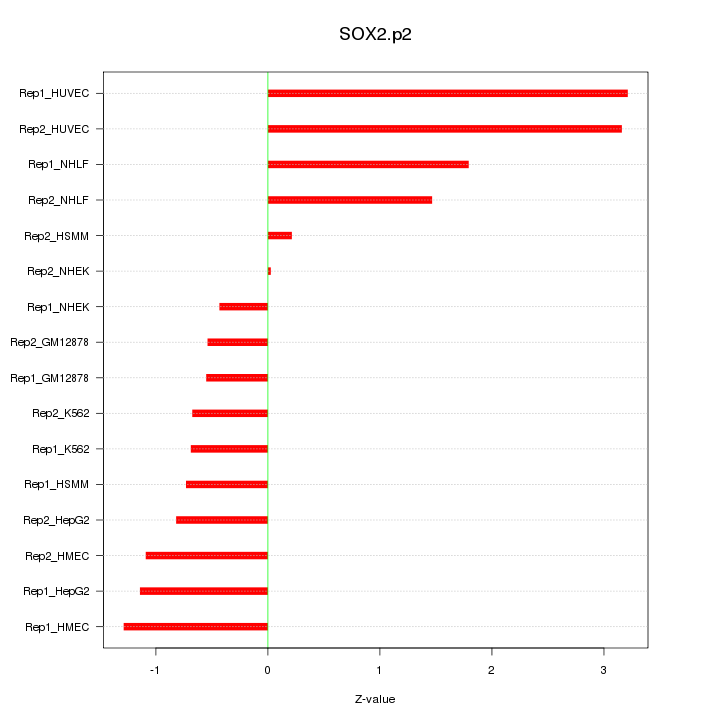
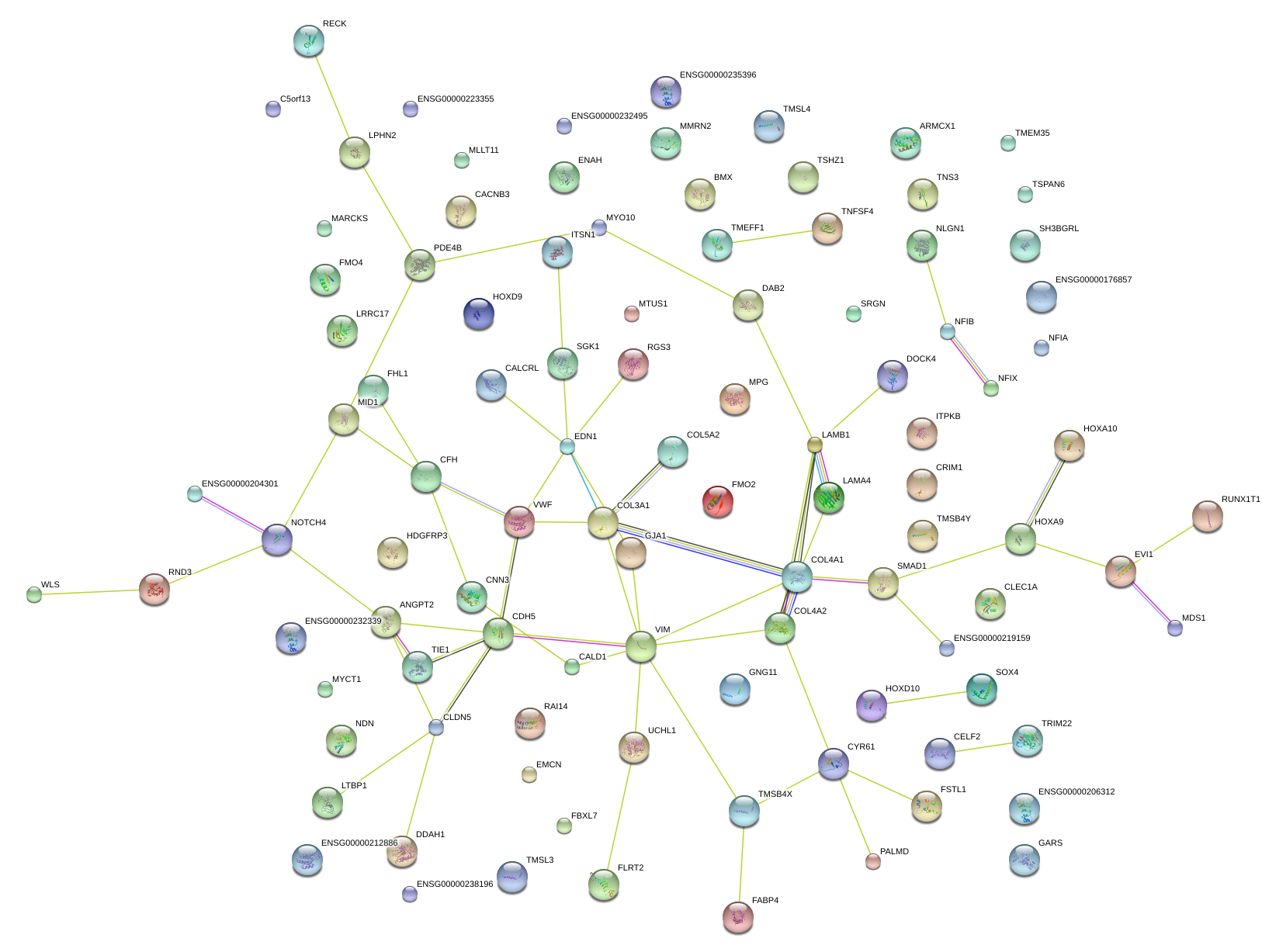
|
chr12_-_6103945
|
3.670
|
NM_000552
|
VWF
|
von Willebrand factor
|
|
chr6_+_12398514
|
3.115
|
NM_001168319
NM_001955
|
EDN1
|
endothelin 1
|
|
chr17_-_59817722
|
3.031
|
NM_000442
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr7_+_102340579
|
3.017
|
NM_001031692
NM_005824
|
LRRC17
|
leucine rich repeat containing 17
|
|
chr2_+_33213111
|
2.891
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr4_-_101658094
|
2.789
|
NM_001159694
NM_016242
|
EMCN
|
endomucin
|
|
chr17_-_59817456
|
2.659
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr17_-_59817575
|
2.656
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr1_+_85819004
|
2.583
|
NM_001554
|
CYR61
|
cysteine-rich, angiogenic inducer, 61
|
|
chr17_-_59817654
|
2.538
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr7_-_111633620
|
2.486
|
NM_014705
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr5_-_39460687
|
2.461
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr2_-_188021117
|
2.452
|
NM_005795
|
CALCRL
|
calcitonin receptor-like
|
|
chr6_+_121798443
|
2.398
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr10_+_70517823
|
2.196
|
NM_002727
|
SRGN
|
serglycin
|
|
chrX_+_15428820
|
2.099
|
NM_203281
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr10_+_70517863
|
2.083
|
|
SRGN
|
serglycin
|
|
chr18_+_71051713
|
2.082
|
NM_005786
|
TSHZ1
|
teashirt zinc finger homeobox 1
|
|
chr6_+_114285209
|
2.049
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr1_-_68470585
|
1.975
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chr4_+_40953698
|
1.946
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr7_-_27171673
|
1.940
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chr4_+_146623405
|
1.835
|
NM_001003688
|
SMAD1
|
SMAD family member 1
|
|
chr7_+_134114957
|
1.820
|
|
CALD1
|
caldesmon 1
|
|
chr7_-_107430566
|
1.780
|
NM_002291
|
LAMB1
|
laminin, beta 1
|
|
chr5_-_111119846
|
1.769
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr1_-_171441234
|
1.764
|
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr7_+_93388951
|
1.748
|
NM_004126
|
GNG11
|
guanine nucleotide binding protein (G protein), gamma 11
|
|
chr8_-_6407882
|
1.736
|
NM_001118887
NM_001118888
NM_001147
|
ANGPT2
|
angiopoietin 2
|
|
chr22_-_17892859
|
1.730
|
NM_001130861
NM_003277
|
CLDN5
|
claudin 5
|
|
chr1_+_81544432
|
1.719
|
|
LPHN2
|
latrophilin 2
|
|
chr11_+_5667494
|
1.696
|
NM_006074
|
TRIM22
|
tripartite motif containing 22
|
|
chr13_-_109757442
|
1.650
|
NM_001845
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr5_-_39461054
|
1.630
|
NM_001343
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr7_-_27186400
|
1.625
|
NM_153715
|
HOXA10
|
homeobox A10
|
|
chr5_+_15553288
|
1.596
|
NM_012304
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chrX_+_100692071
|
1.586
|
NM_016608
|
ARMCX1
|
armadillo repeat containing, X-linked 1
|
|
chr4_+_40953666
|
1.577
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr16_+_64958025
|
1.548
|
NM_001795
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr4_+_40953652
|
1.537
|
NM_004181
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr2_-_151052391
|
1.517
|
NM_005168
|
RND3
|
Rho family GTPase 3
|
|
chrX_+_100220491
|
1.501
|
NM_021637
|
TMEM35
|
transmembrane protein 35
|
|
chr1_-_95165037
|
1.461
|
|
CNN3
|
calponin 3, acidic
|
|
chr8_-_17624009
|
1.453
|
NM_001001931
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr5_-_39460787
|
1.430
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr1_+_66570363
|
1.424
|
NM_001037339
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr2_-_189752575
|
1.398
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr5_+_34723420
|
1.393
|
NM_001145523
|
RAI14
|
retinoic acid induced 14
|
|
chr1_-_95165089
|
1.373
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr10_+_17311282
|
1.371
|
|
VIM
|
vimentin
|
|
chr10_+_17311283
|
1.370
|
|
VIM
|
vimentin
|
|
chr10_+_17311307
|
1.353
|
|
VIM
|
vimentin
|
|
chr3_-_121652183
|
1.342
|
NM_007085
|
FSTL1
|
follistatin-like 1
|
|
chr10_+_17311242
|
1.321
|
|
VIM
|
vimentin
|
|
chr1_+_149299307
|
1.321
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr9_+_115382760
|
1.319
|
NM_021106
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr9_+_115382990
|
1.316
|
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr6_+_153060722
|
1.301
|
NM_025107
|
MYCT1
|
myc target 1
|
|
chr9_+_115303527
|
1.261
|
NM_130795
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr1_-_85703198
|
1.252
|
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr6_-_32271868
|
1.251
|
|
NOTCH4
|
notch 4
|
|
chr6_-_134680888
|
1.238
|
NM_001143676
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr1_+_99884018
|
1.227
|
NM_017734
|
PALMD
|
palmdelphin
|
|
chr10_+_17310475
|
1.219
|
|
VIM
|
vimentin
|
|
chrX_-_10605726
|
1.204
|
NM_001098624
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr12_+_47498778
|
1.201
|
NM_000725
|
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chrX_+_135079461
|
1.196
|
NM_001167819
|
FHL1
|
four and a half LIM domains 1
|
|
chr21_+_34112456
|
1.188
|
|
ITSN1
|
intersectin 1 (SH3 domain protein)
|
|
chr10_+_11246934
|
1.188
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr10_+_11247059
|
1.185
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chrX_+_135079120
|
1.178
|
NM_001159704
|
FHL1
|
four and a half LIM domains 1
|
|
chr7_-_47588266
|
1.177
|
|
TNS3
|
tensin 3
|
|
chrX_+_55951280
|
1.168
|
|
|
|
|
chr9_-_14388981
|
1.161
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr1_-_224993498
|
1.159
|
NM_002221
|
ITPKB
|
inositol 1,4,5-trisphosphate 3-kinase B
|
|
chr1_+_66230977
|
1.158
|
NM_001037340
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr3_+_174598937
|
1.148
|
NM_014932
|
NLGN1
|
neuroligin 1
|
|
chr8_-_93099040
|
1.143
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr9_+_102275310
|
1.135
|
NM_003692
|
TMEFF1
|
transmembrane protein with EGF-like and two follistatin-like domains 1
|
|
chr7_-_47588160
|
1.129
|
|
TNS3
|
tensin 3
|
|
chr3_-_170347019
|
1.128
|
NM_001105078
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr15_-_21483457
|
1.102
|
|
NDN
|
necdin homolog (mouse)
|
|
chr9_+_36026900
|
1.091
|
NM_021111
|
RECK
|
reversion-inducing-cysteine-rich protein with kazal motifs
|
|
chrX_+_80343958
|
1.090
|
NM_003022
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chr10_+_11087259
|
1.087
|
NM_001025077
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr1_+_169421011
|
1.085
|
NM_001460
|
FMO2
|
flavin containing monooxygenase 2 (non-functional)
|
|
chr10_+_11087280
|
1.082
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chrX_+_12904207
|
1.079
|
|
TMSB4X
TMSL3
|
thymosin beta 4, X-linked
thymosin-like 3
|
|
chrX_+_15435351
|
1.079
|
NM_001721
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr12_-_10142779
|
1.078
|
NM_016511
|
CLEC1A
|
C-type lectin domain family 1, member A
|
|
chr6_-_112682573
|
1.066
|
|
LAMA4
|
laminin, alpha 4
|
|
chr5_-_16989371
|
1.059
|
NM_012334
|
MYO10
|
myosin X
|
|
chr2_+_36436873
|
1.057
|
NM_016441
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr6_+_21701942
|
1.044
|
NM_003107
|
SOX4
|
SRY (sex determining region Y)-box 4
|
|
chr2_+_189547377
|
1.042
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr15_-_21483503
|
1.019
|
NM_002487
|
NDN
|
necdin homolog (mouse)
|
|
chr1_-_85816520
|
1.019
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr10_-_88707299
|
1.013
|
NM_024756
|
MMRN2
|
multimerin 2
|
|
chr1_+_43539239
|
1.007
|
NM_005424
|
TIE1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1
|
|
chrX_-_99778340
|
0.997
|
NM_003270
|
TSPAN6
|
tetraspanin 6
|
|
chr15_-_81667773
|
0.995
|
|
HDGFRP3
|
hepatoma-derived growth factor, related protein 3
|
|
chr1_-_223907283
|
0.990
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr8_-_82557956
|
0.980
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chrX_+_80344215
|
0.977
|
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chr13_+_109935250
|
0.971
|
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr9_-_14303899
|
0.971
|
|
NFIB
|
nuclear factor I/B
|
|
chr8_-_93176618
|
0.959
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr14_+_85066207
|
0.958
|
NM_013231
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr1_+_149299530
|
0.944
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chrX_+_12903145
|
0.941
|
NM_021109
NM_183049
|
TMSB4X
TMSL3
|
thymosin beta 4, X-linked
thymosin-like 3
|
|
chrX_+_80344253
|
0.936
|
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chrX_+_135057202
|
0.935
|
NM_001159702
NM_001159703
NM_001449
|
FHL1
|
four and a half LIM domains 1
|
|
chr7_+_16759845
|
0.933
|
NM_014399
|
TSPAN13
|
tetraspanin 13
|
|
chr4_+_91035025
|
0.931
|
NM_007351
|
MMRN1
|
multimerin 1
|
|
chr1_+_149299504
|
0.930
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr1_-_79244948
|
0.927
|
NM_022159
|
ELTD1
|
EGF, latrophilin and seven transmembrane domain containing 1
|
|
chr17_-_3518654
|
0.925
|
NM_014604
|
TAX1BP3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3
|
|
chr13_-_98428244
|
0.923
|
NM_001130048
NM_001130050
|
DOCK9
|
dedicator of cytokinesis 9
|
|
chr17_-_3518568
|
0.917
|
|
TAX1BP3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3
|
|
chr1_+_149299490
|
0.912
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr14_+_76297885
|
0.911
|
NM_014909
|
VASH1
|
vasohibin 1
|
|
chr7_-_130891861
|
0.903
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr1_+_161305188
|
0.898
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr15_+_94670160
|
0.897
|
NM_001145155
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr8_+_70567634
|
0.897
|
|
SULF1
|
sulfatase 1
|
|
chr7_-_27171648
|
0.891
|
|
HOXA9
|
homeobox A9
|
|
chr1_+_154120170
|
0.891
|
|
SYT11
|
synaptotagmin XI
|
|
chr4_+_146622154
|
0.887
|
NM_005900
|
SMAD1
|
SMAD family member 1
|
|
chr5_+_140719773
|
0.885
|
NM_018923
NM_032096
|
PCDHGB2
|
protocadherin gamma subfamily B, 2
|
|
chr4_-_91979113
|
0.885
|
NM_183049
|
TMSL3
|
thymosin-like 3
|
|
chr10_-_29794484
|
0.884
|
|
SVIL
|
supervillin
|
|
chr15_-_81667169
|
0.871
|
NM_016073
|
HDGFRP3
|
hepatoma-derived growth factor, related protein 3
|
|
chr1_+_181871830
|
0.866
|
NM_015149
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr8_+_79591074
|
0.857
|
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr7_-_11838260
|
0.846
|
NM_015204
|
THSD7A
|
thrombospondin, type I, domain containing 7A
|
|
chr7_-_27190882
|
0.843
|
|
HOXA11
|
homeobox A11
|
|
chr3_+_153469421
|
0.842
|
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr8_+_70567581
|
0.838
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr21_-_38792173
|
0.834
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr9_-_14303509
|
0.822
|
|
NFIB
|
nuclear factor I/B
|
|
chr1_+_161305558
|
0.820
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr20_+_11846536
|
0.820
|
NM_014962
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr3_-_150578148
|
0.814
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr7_-_41709191
|
0.811
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chrX_-_37964924
|
0.803
|
NM_001170750
NM_001170751
NM_001170752
NM_006307
|
SRPX
|
sushi-repeat containing protein, X-linked
|
|
chr22_+_36422940
|
0.803
|
NM_001039141
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr5_-_111120929
|
0.796
|
NM_001142482
NM_001142481
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr3_+_161040343
|
0.791
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr9_-_14076901
|
0.786
|
|
NFIB
|
nuclear factor I/B
|
|
chr1_-_199742871
|
0.783
|
NM_001193570
NM_004078
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr1_-_110085182
|
0.780
|
NM_000849
|
GSTM3
|
glutathione S-transferase mu 3 (brain)
|
|
chr15_+_65145248
|
0.772
|
NM_005902
|
SMAD3
|
SMAD family member 3
|
|
chr21_+_36429079
|
0.769
|
NM_001236
|
CBR3
|
carbonyl reductase 3
|
|
chr3_-_124822113
|
0.766
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr8_-_93144366
|
0.764
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr3_+_161040439
|
0.764
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr11_-_6633427
|
0.755
|
NM_003737
|
DCHS1
|
dachsous 1 (Drosophila)
|
|
chr10_+_53744043
|
0.754
|
NM_012242
|
DKK1
|
dickkopf homolog 1 (Xenopus laevis)
|
|
chr3_-_101316038
|
0.754
|
NM_001042459
NM_182909
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr3_+_115099007
|
0.753
|
NM_001172105
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chr3_-_124821868
|
0.746
|
|
MYLK
|
myosin light chain kinase
|
|
chr17_+_16886491
|
0.745
|
NM_015134
NM_201274
|
MPRIP
|
myosin phosphatase Rho interacting protein
|
|
chr17_-_30783631
|
0.740
|
NM_018042
|
SLFN12
|
schlafen family member 12
|
|
chr9_-_87904844
|
0.739
|
NM_177937
|
GOLM1
|
golgi membrane protein 1
|
|
chr12_-_90100679
|
0.736
|
|
DCN
|
decorin
|
|
chr11_+_129496792
|
0.736
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr6_-_128883125
|
0.733
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr4_+_87070449
|
0.730
|
NM_031305
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr5_+_140871107
|
0.725
|
|
|
|
|
chr9_+_115396020
|
0.723
|
NM_144489
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr1_-_199742841
|
0.722
|
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr5_-_111120817
|
0.721
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr18_-_51406359
|
0.720
|
|
TCF4
|
transcription factor 4
|
|
chr17_-_3518628
|
0.715
|
|
TAX1BP3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3
|
|
chr2_-_26058941
|
0.713
|
NM_002254
|
KIF3C
|
kinesin family member 3C
|
|
chr15_-_88148538
|
0.706
|
|
ANPEP
|
alanyl (membrane) aminopeptidase
|
|
chr2_+_188865634
|
0.705
|
NM_016315
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1
|
|
chr2_-_26058831
|
0.704
|
|
KIF3C
|
kinesin family member 3C
|
|
chr5_+_71538456
|
0.703
|
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr14_-_91483757
|
0.703
|
NM_006329
|
FBLN5
|
fibulin 5
|
|
chr6_-_11340887
|
0.699
|
NM_006403
NM_182966
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr12_-_90100710
|
0.696
|
|
DCN
|
decorin
|
|
chr12_+_14429499
|
0.694
|
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr1_-_32574185
|
0.687
|
NM_023009
|
MARCKSL1
|
MARCKS-like 1
|
|
chr5_-_168204430
|
0.685
|
|
|
|
|
chr1_-_26105954
|
0.684
|
NM_203399
|
STMN1
|
stathmin 1
|
|
chr2_-_217248967
|
0.684
|
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr8_-_27524744
|
0.684
|
NM_203339
|
CLU
|
clusterin
|
|
chr14_-_104515738
|
0.683
|
NM_138420
|
AHNAK2
|
AHNAK nucleoprotein 2
|
|
chr6_-_76022565
|
0.680
|
|
TMEM30A
|
transmembrane protein 30A
|
|
chr14_+_105012106
|
0.678
|
NM_001312
|
CRIP2
|
cysteine-rich protein 2
|
|
chr2_+_66516196
|
0.677
|
|
MEIS1
|
Meis homeobox 1
|
|
chr10_-_30064600
|
0.677
|
NM_003174
|
SVIL
|
supervillin
|
|
chr6_-_139737432
|
0.677
|
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr6_-_11340869
|
0.669
|
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr9_+_4480193
|
0.668
|
NM_004170
|
SLC1A1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1
|
|
chr12_-_90100733
|
0.662
|
NM_001920
|
DCN
|
decorin
|
|
chr2_+_188865796
|
0.660
|
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1
|
|
chr7_+_93862137
|
0.660
|
|
COL1A2
PLAG1
|
collagen, type I, alpha 2
pleiomorphic adenoma gene 1
|
|
chr8_-_93184629
|
0.660
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|