|
chr12_+_68028397
|
4.890
|
NM_000239
|
LYZ
|
lysozyme
|
|
chr12_+_68028407
|
4.489
|
|
LYZ
|
lysozyme
|
|
chr1_+_196874759
|
2.864
|
NM_002838
NM_080921
NM_080923
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr1_+_196874867
|
2.776
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr1_+_196874839
|
2.533
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr3_-_150422474
|
2.526
|
NM_000096
|
CP
|
ceruloplasmin (ferroxidase)
|
|
chr11_+_22652894
|
2.301
|
NM_005256
|
GAS2
|
growth arrest-specific 2
|
|
chr6_-_49712483
|
2.010
|
NM_000324
|
RHAG
|
Rh-associated glycoprotein
|
|
chr2_+_228386800
|
1.861
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr9_-_115880494
|
1.804
|
NM_001633
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr4_+_100714995
|
1.581
|
|
MTTP
|
microsomal triglyceride transfer protein
|
|
chr17_-_46479127
|
1.543
|
|
SPAG9
|
sperm associated antigen 9
|
|
chr4_+_71096830
|
1.521
|
NM_017855
|
ODAM
|
odontogenic, ameloblast asssociated
|
|
chr17_+_63252704
|
1.510
|
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr4_+_74491272
|
1.489
|
|
ALB
|
albumin
|
|
chr17_+_63252668
|
1.451
|
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr2_+_143603368
|
1.437
|
NM_018460
|
ARHGAP15
|
Rho GTPase activating protein 15
|
|
chr16_+_68238460
|
1.381
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chrX_-_137649180
|
1.376
|
NM_033642
|
FGF13
|
fibroblast growth factor 13
|
|
chr1_+_116727514
|
1.353
|
NM_001160234
|
ATP1A1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide
|
|
chr2_+_143603439
|
1.350
|
|
ARHGAP15
|
Rho GTPase activating protein 15
|
|
chrX_+_152562013
|
1.347
|
|
DUSP9
|
dual specificity phosphatase 9
|
|
chr6_+_37245860
|
1.280
|
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr9_-_97308687
|
1.240
|
|
PTCH1
|
patched 1
|
|
chr8_-_102872354
|
1.236
|
|
NCALD
|
neurocalcin delta
|
|
chrX_+_133952633
|
1.230
|
NM_001163438
|
LOC644538
|
hypothetical protein LOC644538
|
|
chr2_+_143351536
|
1.202
|
NM_001032998
NM_003937
|
KYNU
|
kynureninase
|
|
chr6_+_27885692
|
1.156
|
NM_003536
|
HIST1H3H
|
histone cluster 1, H3h
|
|
chrY_+_13525412
|
1.149
|
NM_001122665
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr8_-_102872372
|
1.143
|
|
NCALD
|
neurocalcin delta
|
|
chrX_+_123062143
|
1.135
|
|
STAG2
|
stromal antigen 2
|
|
chr12_-_101835183
|
1.123
|
|
PAH
|
phenylalanine hydroxylase
|
|
chr3_-_121884091
|
1.095
|
NM_000187
|
HGD
|
homogentisate 1,2-dioxygenase
|
|
chr12_-_101835510
|
1.094
|
NM_000277
|
PAH
|
phenylalanine hydroxylase
|
|
chr4_+_89115825
|
1.064
|
NM_000582
NM_001040058
NM_001040060
|
SPP1
|
secreted phosphoprotein 1
|
|
chr6_+_64414389
|
1.064
|
NM_015153
|
PHF3
|
PHD finger protein 3
|
|
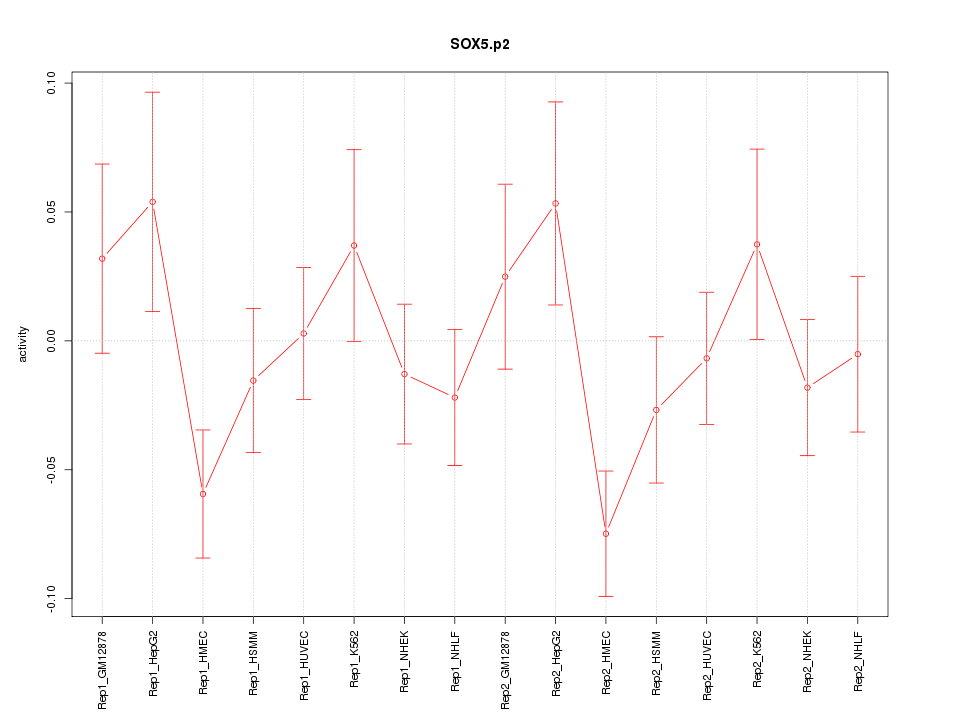
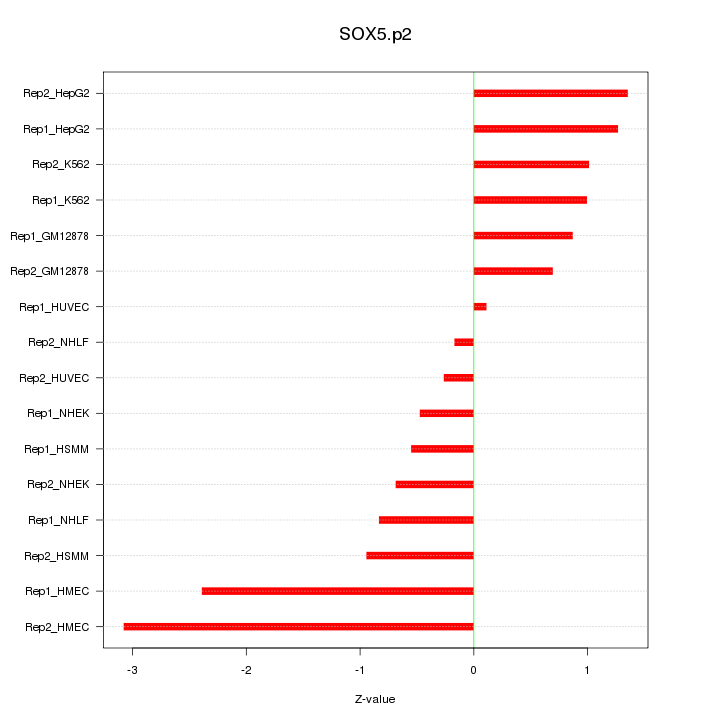
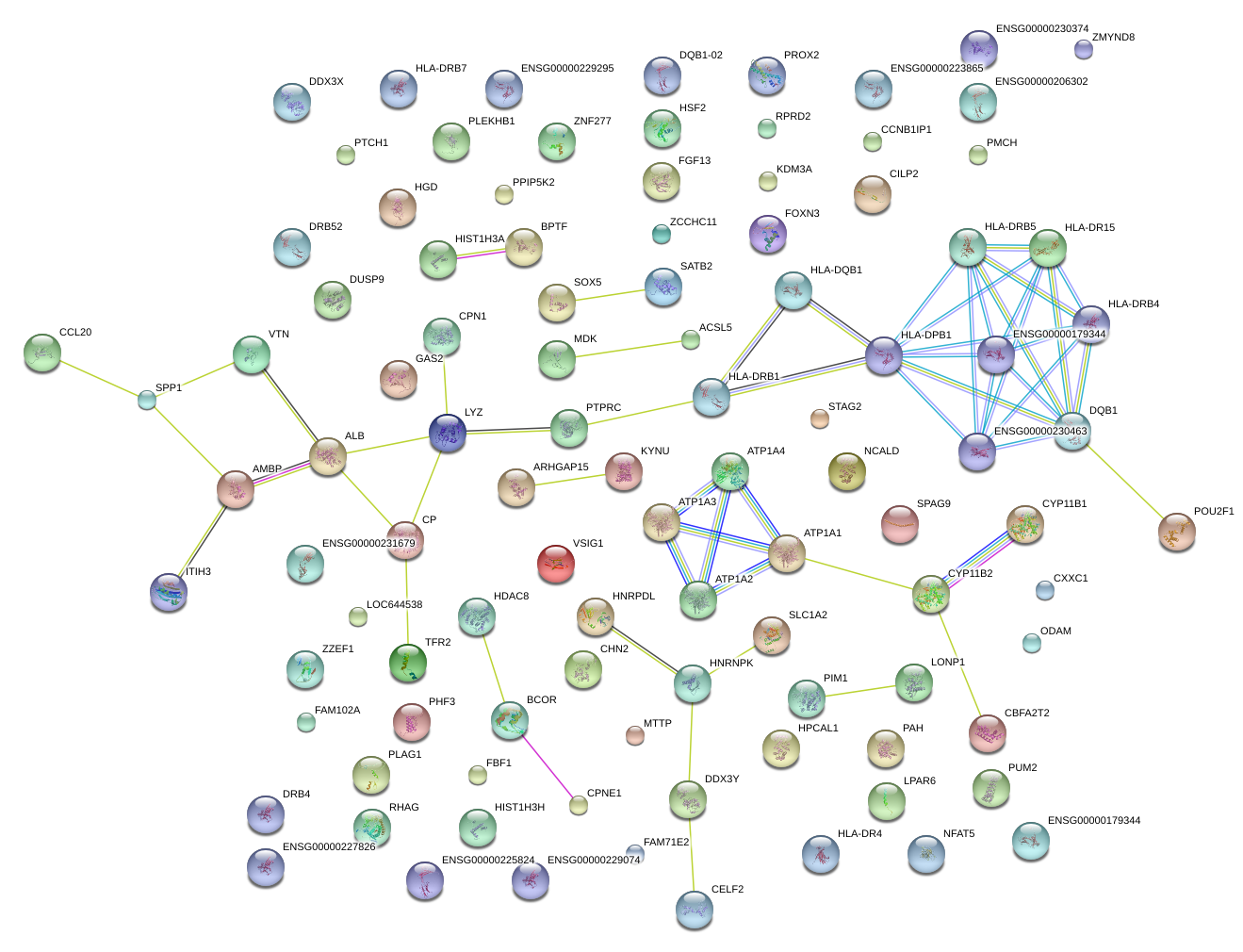
chr12_-_23995220
|
1.062
|
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr20_-_45418818
|
1.060
|
NM_012408
NM_183047
NM_183048
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr12_-_101835026
|
1.043
|
|
PAH
|
phenylalanine hydroxylase
|
|
chr13_-_47885653
|
1.043
|
NM_001162498
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr7_+_111633878
|
1.033
|
NM_021994
|
ZNF277
|
zinc finger protein 277
|
|
chr16_+_68284252
|
1.022
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr12_-_24606616
|
0.986
|
NM_152989
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr11_-_35397090
|
0.982
|
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr9_-_129752687
|
0.980
|
NM_203305
|
FAM102A
|
family with sequence similarity 102, member A
|
|
chr7_-_100077072
|
0.974
|
NM_003227
|
TFR2
|
transferrin receptor 2
|
|
chr6_+_122762384
|
0.969
|
NM_001135564
NM_004506
|
HSF2
|
heat shock transcription factor 2
|
|
chrX_-_39921525
|
0.953
|
NM_001123383
NM_001123384
|
BCOR
|
BCL6 corepressor
|
|
chr17_-_23721420
|
0.952
|
|
VTN
|
vitronectin
|
|
chr11_+_46358909
|
0.949
|
NM_001012334
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr4_+_100704262
|
0.927
|
NM_000253
|
MTTP
|
microsomal triglyceride transfer protein
|
|
chr4_+_89115862
|
0.912
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr17_-_23721351
|
0.898
|
|
VTN
|
vitronectin
|
|
chr20_+_31613800
|
0.893
|
NM_001039709
NM_005093
|
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2
|
|
chr10_-_101831579
|
0.887
|
NM_001308
|
CPN1
|
carboxypeptidase N, polypeptide 1
|
|
chr5_+_102493155
|
0.880
|
NM_015216
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2
|
|
chr10_+_114123828
|
0.843
|
NM_203379
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr7_+_111633972
|
0.838
|
|
ZNF277
|
zinc finger protein 277
|
|
chr4_+_89115892
|
0.823
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr14_-_88953059
|
0.820
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chrX_-_71709335
|
0.811
|
|
HDAC8
|
histone deacetylase 8
|
|
chr11_+_22644732
|
0.807
|
NM_001143830
|
GAS2
|
growth arrest-specific 2
|
|
chr4_-_83566246
|
0.796
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr2_+_86521999
|
0.791
|
NM_001146688
|
KDM3A
|
lysine (K)-specific demethylase 3A
|
|
chr2_-_200033445
|
0.788
|
|
SATB2
|
SATB homeobox 2
|
|
chr10_+_114123765
|
0.777
|
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr10_+_11246934
|
0.774
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr6_+_122762449
|
0.771
|
|
HSF2
|
heat shock transcription factor 2
|
|
chr9_-_85773899
|
0.755
|
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K
|
|
chr3_-_121883647
|
0.753
|
|
HGD
|
homogentisate 1,2-dioxygenase
|
|
chr1_+_148603769
|
0.743
|
|
RPRD2
|
regulation of nuclear pre-mRNA domain containing 2
|
|
chr8_-_57286381
|
0.734
|
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chr11_+_73037382
|
0.734
|
NM_001130036
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr11_-_35397675
|
0.726
|
NM_004171
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr6_-_32665409
|
0.722
|
NM_002124
|
HLA-DRB1
HLA-DRB4
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 1
major histocompatibility complex, class II, DR beta 4
major histocompatibility complex, class II, DR beta 5
|
|
chr1_+_165564904
|
0.719
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr8_-_57286412
|
0.715
|
NM_001114634
NM_001114635
NM_002655
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chr3_+_52803823
|
0.712
|
NM_002217
|
ITIH3
|
inter-alpha (globulin) inhibitor H3
|
|
chr2_-_20375497
|
0.706
|
|
PUM2
|
pumilio homolog 2 (Drosophila)
|
|
chr12_-_101115677
|
0.699
|
NM_002674
|
PMCH
|
pro-melanin-concentrating hormone
|
|
chrX_+_107174855
|
0.683
|
NM_001170553
NM_182607
|
VSIG1
|
V-set and immunoglobulin domain containing 1
|
|
chr14_-_19871267
|
0.676
|
NM_021178
NM_182849
|
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase
|
|
chr2_+_86521939
|
0.669
|
|
KDM3A
|
lysine (K)-specific demethylase 3A
|
|
chr1_-_52791218
|
0.665
|
|
ZCCHC11
|
zinc finger, CCHC domain containing 11
|
|
chr7_+_29486010
|
0.659
|
NM_001039936
|
CHN2
|
chimerin (chimaerin) 2
|
|
chr4_-_155731288
|
0.658
|
NM_000508
NM_021871
|
FGA
|
fibrinogen alpha chain
|
|
chr20_+_254205
|
0.655
|
NM_006943
|
SOX12
|
SRY (sex determining region Y)-box 12
|
|
chr12_-_23995109
|
0.644
|
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr4_-_140280031
|
0.641
|
NM_201999
|
ELF2
|
E74-like factor 2 (ets domain transcription factor)
|
|
chr1_-_52791309
|
0.635
|
NM_001009881
NM_015269
|
ZCCHC11
|
zinc finger, CCHC domain containing 11
|
|
chr22_+_38652555
|
0.633
|
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr1_+_165565006
|
0.633
|
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr1_+_11674365
|
0.632
|
NM_198545
|
C1orf187
|
chromosome 1 open reading frame 187
|
|
chr6_-_79732391
|
0.630
|
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr14_-_106184988
|
0.629
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr7_+_50314801
|
0.620
|
NM_006060
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr14_+_38653177
|
0.618
|
NM_001009182
NM_001009183
NM_003616
|
SIP1
|
survival of motor neuron protein interacting protein 1
|
|
chr4_+_89115923
|
0.613
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr5_-_88155360
|
0.610
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr2_+_74914742
|
0.610
|
|
HK2
|
hexokinase 2
|
|
chr3_+_46591048
|
0.608
|
NM_001174136
|
TDGF1
|
teratocarcinoma-derived growth factor 1
|
|
chr6_-_43135092
|
0.607
|
NM_015950
|
MRPL2
|
mitochondrial ribosomal protein L2
|
|
chr6_+_88026338
|
0.605
|
|
ZNF292
|
zinc finger protein 292
|
|
chr6_-_32605898
|
0.603
|
NM_002125
|
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 5
|
|
chr4_-_71751070
|
0.599
|
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides
|
|
chr2_-_20390601
|
0.596
|
NM_015317
|
PUM2
|
pumilio homolog 2 (Drosophila)
|
|
chr8_-_102872614
|
0.596
|
NM_032041
|
NCALD
|
neurocalcin delta
|
|
chr9_+_136358145
|
0.582
|
|
RXRA
|
retinoid X receptor, alpha
|
|
chr17_-_44058612
|
0.581
|
NM_024017
|
HOXB9
|
homeobox B9
|
|
chr17_+_56032271
|
0.581
|
NM_003620
|
PPM1D
|
protein phosphatase, Mg2+/Mn2+ dependent, 1D
|
|
chr2_-_162638875
|
0.573
|
|
DPP4
|
dipeptidyl-peptidase 4
|
|
chr9_-_115877104
|
0.571
|
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr4_-_84254873
|
0.570
|
|
PLAC8
|
placenta-specific 8
|
|
chr17_-_36346240
|
0.569
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr12_-_101396459
|
0.567
|
NM_001111284
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr15_+_72861767
|
0.567
|
|
CSK
|
c-src tyrosine kinase
|
|
chr9_-_122728173
|
0.566
|
|
TRAF1
|
TNF receptor-associated factor 1
|
|
chr4_-_105635499
|
0.566
|
|
CXXC4
|
CXXC finger protein 4
|
|
chr4_+_124537334
|
0.564
|
NM_199327
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr17_-_77488258
|
0.553
|
NM_006907
NM_153824
|
PYCR1
|
pyrroline-5-carboxylate reductase 1
|
|
chr10_-_99084417
|
0.550
|
NM_012083
|
FRAT2
|
frequently rearranged in advanced T-cell lymphomas 2
|
|
chr15_+_72861470
|
0.550
|
NM_001127190
NM_004383
|
CSK
|
c-src tyrosine kinase
|
|
chr1_-_51660328
|
0.549
|
NM_001159969
|
EPS15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr11_-_101828563
|
0.549
|
NM_052932
|
TMEM123
|
transmembrane protein 123
|
|
chrX_+_49047068
|
0.542
|
NM_001098413
|
GAGE10
|
G antigen 10
|
|
chr6_-_138862273
|
0.539
|
|
|
|
|
chr2_+_86800898
|
0.538
|
NM_022780
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae)
|
|
chr11_-_72530605
|
0.536
|
NM_014824
|
FCHSD2
|
FCH and double SH3 domains 2
|
|
chr3_-_57209319
|
0.534
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chr4_-_71751205
|
0.531
|
NM_144646
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides
|
|
chr14_-_90953883
|
0.527
|
NM_001080414
|
CCDC88C
|
coiled-coil domain containing 88C
|
|
chr1_-_74971679
|
0.525
|
NM_001130042
NM_001130043
NM_001134759
NM_001889
|
CRYZ
|
crystallin, zeta (quinone reductase)
|
|
chr1_-_229627412
|
0.523
|
NM_022051
|
EGLN1
|
egl nine homolog 1 (C. elegans)
|
|
chr2_+_86521781
|
0.522
|
NM_018433
|
KDM3A
|
lysine (K)-specific demethylase 3A
|
|
chr4_+_170817787
|
0.521
|
|
CLCN3
|
chloride channel 3
|
|
chr1_+_50344550
|
0.520
|
NM_001144775
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chrX_+_9392980
|
0.520
|
NM_005647
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr12_-_110410730
|
0.516
|
|
ATXN2
|
ataxin 2
|
|
chr5_-_71838875
|
0.508
|
NM_152625
|
ZNF366
|
zinc finger protein 366
|
|
chr2_-_162639297
|
0.507
|
NM_001935
|
DPP4
|
dipeptidyl-peptidase 4
|
|
chr1_-_180627023
|
0.505
|
NM_001033056
|
GLUL
|
glutamate-ammonia ligase
|
|
chr4_-_187114407
|
0.503
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr5_-_175896829
|
0.501
|
NM_014901
|
RNF44
|
ring finger protein 44
|
|
chr2_-_174475528
|
0.498
|
|
|
|
|
chr2_+_86800940
|
0.492
|
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae)
|
|
chr2_+_232281470
|
0.491
|
NM_001099285
NM_002823
|
PTMA
|
prothymosin, alpha
|
|
chr6_+_17708483
|
0.491
|
NM_016255
|
FAM8A1
|
family with sequence similarity 8, member A1
|
|
chr16_-_45562840
|
0.490
|
|
DNAJA2
|
DnaJ (Hsp40) homolog, subfamily A, member 2
|
|
chr16_+_2593351
|
0.489
|
|
LOC652276
|
hypothetical LOC652276
|
|
chr19_+_3958690
|
0.488
|
NM_015897
|
PIAS4
|
protein inhibitor of activated STAT, 4
|
|
chr1_-_148508030
|
0.484
|
NM_001077628
NM_016022
|
APH1A
|
anterior pharynx defective 1 homolog A (C. elegans)
|
|
chr5_-_32424352
|
0.481
|
|
ZFR
|
zinc finger RNA binding protein
|
|
chr17_+_65009986
|
0.476
|
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr7_-_16471817
|
0.475
|
NM_015464
|
SOSTDC1
|
sclerostin domain containing 1
|
|
chr19_-_46595053
|
0.474
|
NM_020158
|
EXOSC5
|
exosome component 5
|
|
chr5_+_36912656
|
0.472
|
|
NIPBL
|
Nipped-B homolog (Drosophila)
|
|
chr1_-_180626804
|
0.470
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr9_-_74757735
|
0.469
|
NM_000689
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr17_-_59817722
|
0.467
|
NM_000442
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr4_-_73653331
|
0.467
|
NM_014243
|
ADAMTS3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3
|
|
chr17_+_56032385
|
0.464
|
|
PPM1D
|
protein phosphatase, Mg2+/Mn2+ dependent, 1D
|
|
chr3_-_47909001
|
0.463
|
|
MAP4
|
microtubule-associated protein 4
|
|
chr1_-_12599934
|
0.462
|
NM_004753
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3
|
|
chr2_+_232281491
|
0.461
|
|
PTMA
|
prothymosin, alpha
|
|
chrX_+_70419766
|
0.456
|
NM_001145408
NM_001145409
NM_001145410
NM_007363
|
NONO
|
non-POU domain containing, octamer-binding
|
|
chr10_+_101532450
|
0.456
|
NM_000392
|
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2
|
|
chr13_-_47885248
|
0.453
|
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr7_+_116447499
|
0.453
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr4_+_74500748
|
0.450
|
|
ALB
|
albumin
|
|
chr13_-_45577144
|
0.450
|
NM_001872
NM_016413
|
CPB2
|
carboxypeptidase B2 (plasma)
|
|
chr15_+_80741876
|
0.448
|
|
LOC80154
|
hypothetical LOC80154
|
|
chr3_+_173043832
|
0.446
|
NM_001164436
|
TMEM212
|
transmembrane protein 212
|
|
chr1_-_148507779
|
0.445
|
|
APH1A
|
anterior pharynx defective 1 homolog A (C. elegans)
|
|
chr20_-_45410033
|
0.444
|
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr6_+_32090516
|
0.443
|
NM_007293
NM_001002029
|
C4A
C4B
|
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr1_-_31003181
|
0.442
|
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chr12_-_31635089
|
0.442
|
NM_144973
|
DENND5B
|
DENN/MADD domain containing 5B
|
|
chr4_-_84254932
|
0.441
|
NM_001130715
NM_016619
|
PLAC8
|
placenta-specific 8
|
|
chr7_+_96583840
|
0.440
|
NM_020186
|
ACN9
|
ACN9 homolog (S. cerevisiae)
|
|
chr19_-_51909416
|
0.440
|
NM_001079882
|
PRKD2
|
protein kinase D2
|
|
chr19_-_52426055
|
0.439
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr18_+_27425727
|
0.438
|
NM_000371
|
TTR
|
transthyretin
|
|
chr8_-_70907957
|
0.435
|
NM_001146009
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1
|
|
chr6_+_155579788
|
0.434
|
NM_001010927
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2
|
|
chr17_-_36074812
|
0.434
|
NM_152349
|
KRT222
|
keratin 222
|
|
chr10_+_35455986
|
0.434
|
|
CREM
|
cAMP responsive element modulator
|
|
chr12_-_101398451
|
0.433
|
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr3_-_12916547
|
0.433
|
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr4_+_69996780
|
0.431
|
NM_001074
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7
|
|
chr10_+_35455806
|
0.427
|
NM_001881
NM_182850
NM_182853
NM_183013
|
CREM
|
cAMP responsive element modulator
|
|
chrX_-_78509662
|
0.427
|
NM_001171581
NM_004867
|
ITM2A
|
integral membrane protein 2A
|
|
chr1_+_50347188
|
0.425
|
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr8_-_108579270
|
0.424
|
NM_001146
|
ANGPT1
|
angiopoietin 1
|
|
chr8_+_126511739
|
0.422
|
NM_025195
|
TRIB1
|
tribbles homolog 1 (Drosophila)
|
|
chr3_-_143230124
|
0.414
|
NM_001178141
NM_001178142
NM_006286
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr16_-_66535457
|
0.412
|
NM_000229
|
LCAT
|
lecithin-cholesterol acyltransferase
|
|
chr16_-_30841938
|
0.411
|
|
NCRNA00095
|
non-protein coding RNA 95
|
|
chr8_-_70908130
|
0.411
|
NM_001146008
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1
|
|
chr10_-_5050191
|
0.410
|
NM_001354
NM_205845
|
AKR1C2
|
aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III)
|
|
chr17_-_53847937
|
0.409
|
|
RNF43
|
ring finger protein 43
|