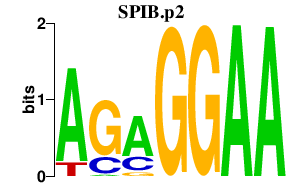
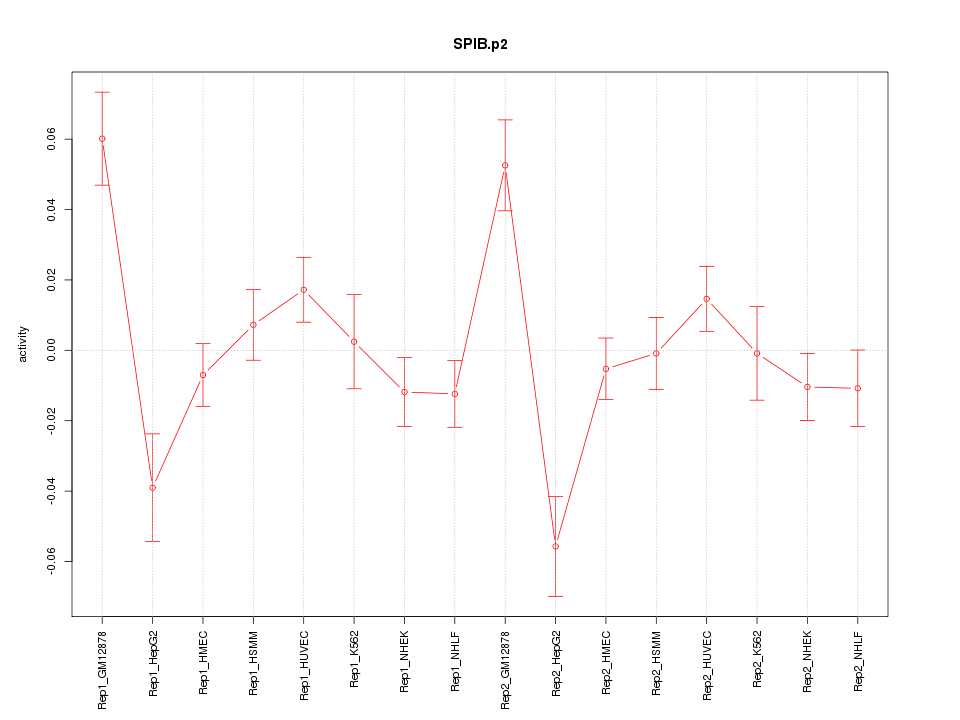
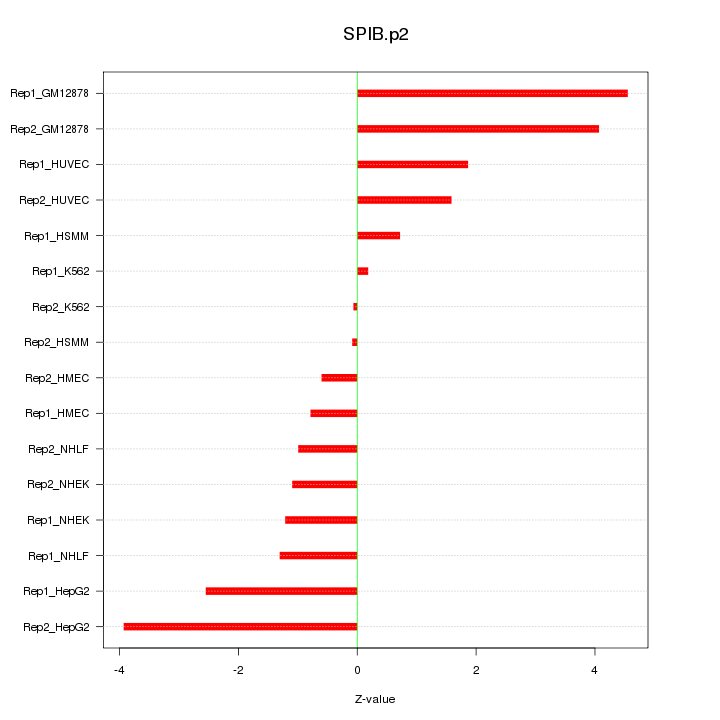
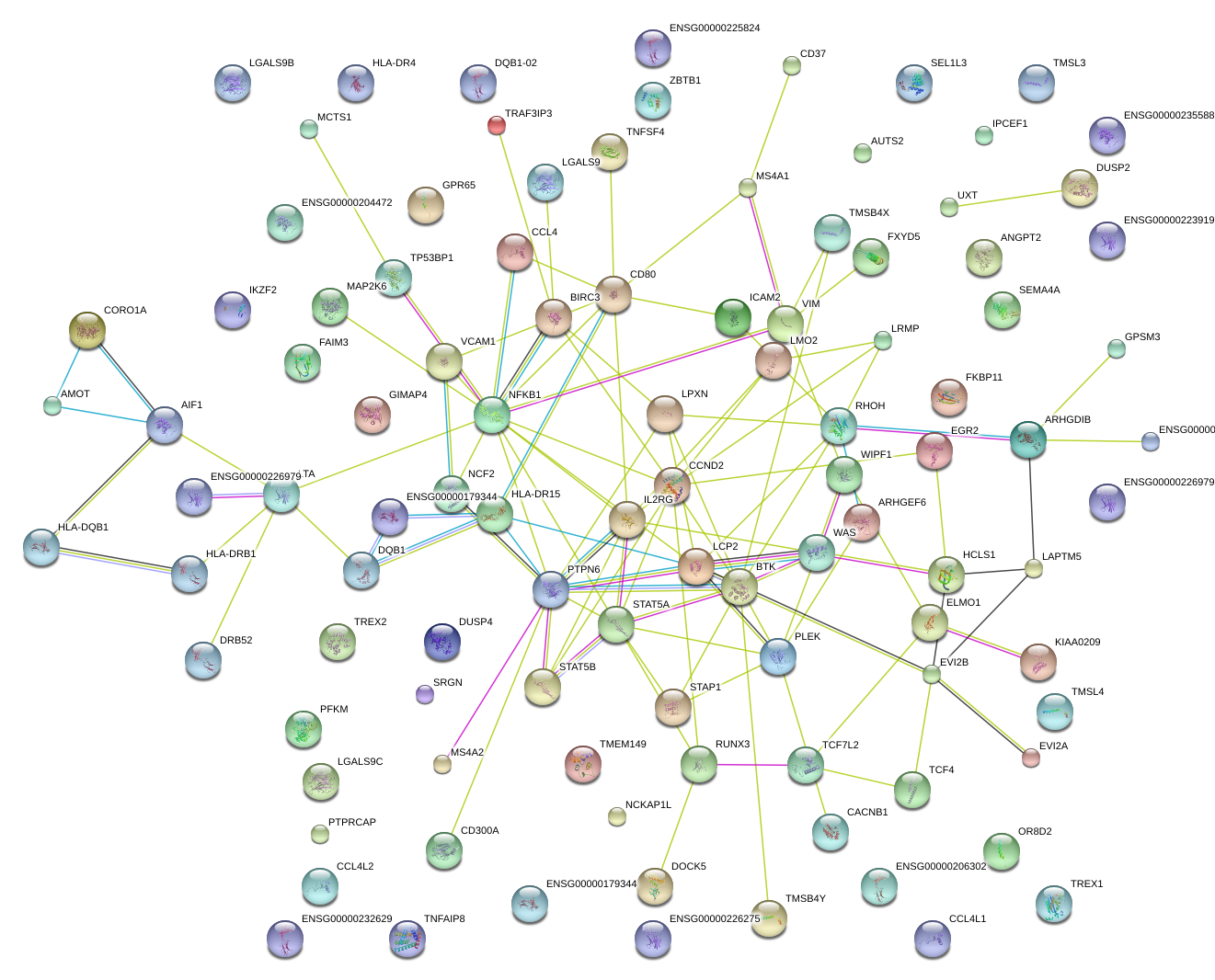
|
chr12_-_15005761
|
8.931
|
NM_001175
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta
|
|
chrX_+_48427103
|
4.987
|
NM_000377
|
WAS
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia)
|
|
chr18_-_51219880
|
4.463
|
|
TCF4
|
transcription factor 4
|
|
chr16_+_30102426
|
3.259
|
|
CORO1A
|
coronin, actin binding protein, 1A
|
|
chr2_-_175207521
|
3.249
|
NM_003387
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr6_-_32742265
|
3.113
|
NM_002123
|
HLA-DQB1
|
major histocompatibility complex, class II, DQ beta 1
|
|
chr10_-_64246129
|
3.112
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr17_+_31455332
|
2.897
|
NM_002984
|
CCL4
|
chemokine (C-C motif) ligand 4
|
|
chr12_+_25096755
|
2.856
|
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr2_-_96174865
|
2.693
|
NM_004418
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr17_+_31664143
|
2.613
|
NM_207007
NM_001001435
|
CCL4L2
CCL4L1
|
chemokine (C-C motif) ligand 4-like 2
chemokine (C-C motif) ligand 4-like 1
|
|
chr3_-_122862447
|
2.577
|
NM_005335
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr22_+_21552897
|
2.473
|
|
LOC100290481
|
immunoglobulin lambda light chain-like
|
|
chr19_+_54530497
|
2.446
|
|
CD37
|
CD37 molecule
|
|
chr10_+_70517863
|
2.440
|
|
SRGN
|
serglycin
|
|
chr19_+_54530485
|
2.434
|
NM_001040031
NM_001774
|
CD37
|
CD37 molecule
|
|
chr1_-_181826086
|
2.431
|
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr6_-_32268267
|
2.420
|
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr12_+_25096472
|
2.416
|
NM_006152
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr1_-_181826633
|
2.415
|
NM_001127651
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr10_+_70517823
|
2.411
|
NM_002727
|
SRGN
|
serglycin
|
|
chr19_+_54530512
|
2.365
|
|
CD37
|
CD37 molecule
|
|
chr1_-_25128951
|
2.352
|
|
RUNX3
|
runt-related transcription factor 3
|
|
chr19_+_54530546
|
2.337
|
|
CD37
|
CD37 molecule
|
|
chr3_-_122862391
|
2.317
|
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr19_+_54530458
|
2.268
|
|
CD37
|
CD37 molecule
|
|
chr1_-_181826292
|
2.217
|
NM_000433
NM_001190789
NM_001190794
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr17_+_64922420
|
2.169
|
NM_002758
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr11_+_59979857
|
2.145
|
NM_021950
NM_152866
|
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1
|
|
chr4_+_39874921
|
2.125
|
NM_004310
|
RHOH
|
ras homolog gene family, member H
|
|
chr14_+_87541220
|
2.066
|
NM_003608
|
GPR65
|
G protein-coupled receptor 65
|
|
chr12_+_53177806
|
2.037
|
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr3_+_48482213
|
2.020
|
NM_016381
NM_033629
|
TREX1
|
three prime repair exonuclease 1
|
|
chr17_-_26665220
|
1.975
|
NM_006495
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr12_+_46785922
|
1.968
|
NM_001166686
|
PFKM
|
phosphofructokinase, muscle
|
|
chr12_+_4253143
|
1.925
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr17_-_26665213
|
1.924
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chrX_+_48427137
|
1.909
|
|
WAS
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia)
|
|
chr12_+_53177789
|
1.894
|
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr7_+_68701704
|
1.881
|
NM_001127231
NM_001127232
NM_015570
|
AUTS2
|
autism susceptibility candidate 2
|
|
chr17_+_31562577
|
1.850
|
NM_207007
NM_001001435
|
CCL4L2
CCL4L1
|
chemokine (C-C motif) ligand 4-like 2
chemokine (C-C motif) ligand 4-like 1
|
|
chr12_+_53177758
|
1.846
|
NM_005337
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr22_+_21495273
|
1.843
|
|
|
|
|
chr5_-_169657312
|
1.786
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr17_-_26665144
|
1.779
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr12_+_6930778
|
1.774
|
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chrX_-_70248022
|
1.769
|
NM_000206
|
IL2RG
|
interleukin 2 receptor, gamma
|
|
chr12_+_6930799
|
1.769
|
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr5_-_169657395
|
1.756
|
NM_005565
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr4_-_25473528
|
1.754
|
NM_015187
|
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans)
|
|
chr12_+_6930792
|
1.753
|
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr4_+_103641854
|
1.751
|
|
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1
|
|
chr12_+_6930804
|
1.744
|
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr6_-_32268615
|
1.704
|
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr5_-_169657360
|
1.667
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr19_-_40925151
|
1.665
|
NM_024660
|
TMEM149
|
transmembrane protein 149
|
|
chr17_-_26665194
|
1.664
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr11_-_58099884
|
1.656
|
NM_004811
|
LPXN
|
leupaxin
|
|
chr17_-_26672757
|
1.652
|
NM_001003927
NM_014210
|
EVI2A
|
ecotropic viral integration site 2A
|
|
chr10_+_17312654
|
1.629
|
|
VIM
|
vimentin
|
|
chrX_-_111953009
|
1.628
|
NM_001113490
|
AMOT
|
angiomotin
|
|
chr11_+_101693403
|
1.622
|
NM_001165
NM_182962
|
BIRC3
|
baculoviral IAP repeat containing 3
|
|
chr1_-_205161821
|
1.618
|
NM_001193338
NM_001142473
NM_005449
|
FAIM3
|
Fas apoptotic inhibitory molecule 3
|
|
chrX_-_135690770
|
1.616
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr12_+_6930694
|
1.613
|
NM_002831
NM_080549
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr11_-_33847937
|
1.609
|
NM_001142316
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr4_+_68107039
|
1.600
|
NM_012108
|
STAP1
|
signal transducing adaptor family member 1
|
|
chr5_+_168996852
|
1.595
|
|
DOCK2
|
dedicator of cytokinesis 2
|
|
chr12_+_6930815
|
1.592
|
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr11_-_33847834
|
1.591
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chrX_-_135690699
|
1.583
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr17_-_59437782
|
1.576
|
NM_000873
|
ICAM2
|
intercellular adhesion molecule 2
|
|
chrX_-_100527799
|
1.542
|
NM_000061
|
BTK
|
Bruton agammaglobulinemia tyrosine kinase
|
|
chr3_-_120761092
|
1.526
|
NM_005191
|
CD80
|
CD80 molecule
|
|
chr7_+_68702252
|
1.490
|
|
AUTS2
|
autism susceptibility candidate 2
|
|
chr1_-_25164061
|
1.471
|
NM_001031680
|
RUNX3
|
runt-related transcription factor 3
|
|
chr11_-_33847507
|
1.429
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr17_+_37693090
|
1.426
|
NM_003152
|
STAT5A
|
signal transducer and activator of transcription 5A
|
|
chr14_-_105762742
|
1.393
|
|
|
|
|
chr7_-_37454933
|
1.374
|
NM_014800
|
ELMO1
|
engulfment and cell motility 1
|
|
chr1_+_207995999
|
1.374
|
NM_025228
|
TRAF3IP3
|
TRAF3 interacting protein 3
|
|
chr1_+_154390939
|
1.361
|
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr1_-_31003260
|
1.353
|
NM_006762
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chr1_-_171441234
|
1.335
|
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr6_-_154719560
|
1.317
|
NM_001130700
NM_015553
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1
|
|
chr1_-_31003181
|
1.306
|
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chr10_-_64246080
|
1.302
|
|
EGR2
|
early growth response 2
|
|
chr17_+_69974361
|
1.301
|
|
CD300A
|
CD300a molecule
|
|
chr6_+_31647854
|
1.292
|
NM_001159740
|
LTA
|
lymphotoxin alpha (TNF superfamily, member 1)
|
|
chr4_+_103641762
|
1.287
|
|
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1
|
|
chr1_+_100957883
|
1.252
|
NM_001078
NM_080682
|
VCAM1
|
vascular cell adhesion molecule 1
|
|
chr8_-_6407882
|
1.240
|
NM_001118887
NM_001118888
NM_001147
|
ANGPT2
|
angiopoietin 2
|
|
chrX_+_119622012
|
1.232
|
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr14_+_64041257
|
1.215
|
|
ZBTB1
|
zinc finger and BTB domain containing 1
|
|
chr2_+_68445821
|
1.207
|
NM_002664
|
PLEK
|
pleckstrin
|
|
chrX_+_12904207
|
1.204
|
|
TMSB4X
TMSL3
|
thymosin beta 4, X-linked
thymosin-like 3
|
|
chr8_-_29263714
|
1.202
|
NM_001394
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr11_-_33848061
|
1.178
|
NM_001142315
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr2_-_213723298
|
1.175
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr5_+_118719591
|
1.163
|
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8
|
|
chr7_+_149895390
|
1.151
|
NM_018326
|
GIMAP4
|
GTPase, IMAP family member 4
|
|
chr19_+_40337846
|
1.142
|
|
FXYD5
|
FXYD domain containing ion transport regulator 5
|
|
chr11_-_66961709
|
1.125
|
NM_005608
|
PTPRCAP
|
protein tyrosine phosphatase, receptor type, C-associated protein
|
|
chr15_-_41572558
|
1.122
|
NM_001141979
NM_001141980
|
TP53BP1
|
tumor protein p53 binding protein 1
|
|
chr5_-_169657192
|
1.122
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr6_+_31690957
|
1.121
|
NM_004847
NM_001623
|
AIF1
|
allograft inflammatory factor 1
|
|
chr17_+_22982372
|
1.088
|
|
LGALS9
|
lectin, galactoside-binding, soluble, 9
|
|
chr19_-_6432765
|
1.085
|
NM_024898
|
DENND1C
|
DENN/MADD domain containing 1C
|
|
chr1_-_25129354
|
1.081
|
NM_004350
|
RUNX3
|
runt-related transcription factor 3
|
|
chr6_-_6265896
|
1.081
|
NM_000129
|
F13A1
|
coagulation factor XIII, A1 polypeptide
|
|
chr4_+_91035025
|
1.064
|
NM_007351
|
MMRN1
|
multimerin 1
|
|
chr11_+_5602782
|
1.064
|
NM_021616
|
TRIM34
|
tripartite motif containing 34
|
|
chr17_+_22982344
|
1.064
|
|
LGALS9
|
lectin, galactoside-binding, soluble, 9
|
|
chr16_+_80370426
|
1.062
|
NM_002661
|
PLCG2
|
phospholipase C, gamma 2 (phosphatidylinositol-specific)
|
|
chr1_+_182040837
|
1.062
|
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr17_+_22982319
|
1.061
|
|
LGALS9
|
lectin, galactoside-binding, soluble, 9
|
|
chr3_-_16530132
|
1.053
|
NM_015150
|
RFTN1
|
raftlin, lipid raft linker 1
|
|
chr11_-_60475558
|
1.052
|
NM_016582
|
SLC15A3
|
solute carrier family 15, member 3
|
|
chr17_+_22982383
|
1.045
|
|
LGALS9
|
lectin, galactoside-binding, soluble, 9
|
|
chr15_+_79376295
|
1.045
|
NM_004513
|
IL16
|
interleukin 16 (lymphocyte chemoattractant factor)
|
|
chr1_-_23758871
|
1.044
|
NM_002167
|
ID3
|
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein
|
|
chr3_+_69216747
|
1.036
|
NM_006407
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5
|
|
chr19_+_40337454
|
1.032
|
NM_014164
|
FXYD5
|
FXYD domain containing ion transport regulator 5
|
|
chr6_+_116707975
|
1.026
|
NM_001080976
|
LOC100506515
DSE
|
hypothetical LOC100506515
dermatan sulfate epimerase
|
|
chr6_+_32515596
|
1.019
|
NM_019111
|
HLA-DRA
|
major histocompatibility complex, class II, DR alpha
|
|
chr17_+_73676219
|
1.018
|
NM_004710
|
SYNGR2
|
synaptogyrin 2
|
|
chrX_-_15782842
|
1.009
|
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr18_-_59137488
|
1.001
|
NM_000633
NM_000657
|
BCL2
|
B-cell CLL/lymphoma 2
|
|
chrX_+_37524190
|
0.992
|
NM_000397
|
CYBB
|
cytochrome b-245, beta polypeptide
|
|
chr11_-_33848013
|
0.991
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr17_+_18320822
|
0.987
|
NM_001040078
|
LGALS9B
LGALS9C
|
lectin, galactoside-binding, soluble, 9B
lectin, galactoside-binding, soluble, 9C
|
|
chr10_-_53129315
|
0.987
|
NM_015235
|
CSTF2T
|
cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa, tau variant
|
|
chr13_-_45654327
|
0.986
|
NM_002298
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr14_-_105206106
|
0.986
|
|
|
|
|
chr19_+_40337480
|
0.983
|
NM_144779
NM_001164605
|
FXYD5
|
FXYD domain containing ion transport regulator 5
|
|
chrX_-_15782826
|
0.979
|
|
|
|
|
chr17_-_35975227
|
0.978
|
NM_001838
|
CCR7
|
chemokine (C-C motif) receptor 7
|
|
chr12_-_101398288
|
0.967
|
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr14_-_105644300
|
0.966
|
|
IGHV4-31
IGHA1
IGHA2
IGHG1
IGHG3
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
|
|
chr22_+_21053972
|
0.966
|
|
|
|
|
chrX_-_15783015
|
0.960
|
NM_003916
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr12_+_53178834
|
0.960
|
NM_001184976
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr17_+_22982278
|
0.953
|
NM_002308
NM_009587
|
LGALS9
|
lectin, galactoside-binding, soluble, 9
|
|
chr12_-_108918440
|
0.951
|
|
GIT2
|
G protein-coupled receptor kinase interacting ArfGAP 2
|
|
chr6_+_31651319
|
0.943
|
NM_000594
|
TNF
|
tumor necrosis factor
|
|
chr19_-_44518516
|
0.943
|
NM_004877
|
GMFG
|
glia maturation factor, gamma
|
|
chrX_-_129072333
|
0.939
|
NM_001127197
|
ELF4
|
E74-like factor 4 (ets domain transcription factor)
|
|
chr8_+_98772537
|
0.930
|
|
MTDH
|
metadherin
|
|
chr8_-_28803341
|
0.925
|
NM_001172562
|
INTS9
|
integrator complex subunit 9
|
|
chr10_+_35524060
|
0.920
|
NM_182717
NM_182718
NM_182719
NM_182720
|
CREM
|
cAMP responsive element modulator
|
|
chr22_-_35875890
|
0.916
|
NM_000878
|
IL2RB
|
interleukin 2 receptor, beta
|
|
chr11_-_10786610
|
0.912
|
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2
|
|
chr10_-_27189898
|
0.910
|
|
ABI1
|
abl-interactor 1
|
|
chr3_-_16529996
|
0.909
|
|
RFTN1
|
raftlin, lipid raft linker 1
|
|
chr16_+_80370400
|
0.907
|
|
PLCG2
|
phospholipase C, gamma 2 (phosphatidylinositol-specific)
|
|
chr4_-_105631915
|
0.899
|
NM_025212
|
CXXC4
|
CXXC finger protein 4
|
|
chr10_+_35524835
|
0.896
|
NM_182721
NM_182722
NM_182723
NM_182724
NM_182725
|
CREM
|
cAMP responsive element modulator
|
|
chr3_+_48482625
|
0.895
|
|
TREX1
|
three prime repair exonuclease 1
|
|
chr6_-_6265874
|
0.895
|
|
F13A1
|
coagulation factor XIII, A1 polypeptide
|
|
chr7_+_128367961
|
0.895
|
NM_001098627
|
IRF5
|
interferon regulatory factor 5
|
|
chr1_-_158883666
|
0.894
|
NM_003037
|
SLAMF1
|
signaling lymphocytic activation molecule family member 1
|
|
chr19_-_44518465
|
0.892
|
|
GMFG
|
glia maturation factor, gamma
|
|
chr22_-_35970230
|
0.892
|
NM_002872
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr12_-_101398503
|
0.887
|
NM_000618
NM_001111283
NM_001111285
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr1_+_26517032
|
0.886
|
|
CD52
|
CD52 molecule
|
|
chr20_+_44180296
|
0.878
|
NM_001250
NM_152854
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5
|
|
chr5_+_168996816
|
0.872
|
NM_004946
|
DOCK2
|
dedicator of cytokinesis 2
|
|
chr12_-_91063441
|
0.872
|
NM_001731
|
BTG1
|
B-cell translocation gene 1, anti-proliferative
|
|
chr1_+_26517045
|
0.869
|
|
CD52
|
CD52 molecule
|
|
chr16_+_30102231
|
0.869
|
NM_001193333
NM_007074
|
CORO1A
|
coronin, actin binding protein, 1A
|
|
chr1_-_23758503
|
0.865
|
|
ID3
|
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein
|
|
chr20_+_29598848
|
0.865
|
|
HM13
|
histocompatibility (minor) 13
|
|
chr1_-_111544784
|
0.860
|
NM_024901
|
DENND2D
|
DENN/MADD domain containing 2D
|
|
chr9_+_115382990
|
0.854
|
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr14_+_49157150
|
0.853
|
NM_002408
|
MGAT2
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase
|
|
chr1_-_111544820
|
0.850
|
|
DENND2D
|
DENN/MADD domain containing 2D
|
|
chr22_-_35970133
|
0.836
|
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr13_-_48008118
|
0.834
|
|
RCBTB2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2
|
|
chr3_+_9809776
|
0.834
|
NM_001198780
|
ARPC4
|
actin related protein 2/3 complex, subunit 4, 20kDa
|
|
chr10_-_98021144
|
0.832
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr1_+_157246305
|
0.831
|
NM_005531
|
IFI16
|
interferon, gamma-inducible protein 16
|
|
chr16_+_30391460
|
0.824
|
NM_001114380
NM_002209
|
ITGAL
|
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide)
|
|
chr1_+_148511806
|
0.821
|
NM_024579
|
C1orf54
|
chromosome 1 open reading frame 54
|
|
chrX_+_119621771
|
0.812
|
NM_014060
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr19_-_40928131
|
0.810
|
NM_001040425
NM_144987
|
U2AF1L4
|
U2 small nuclear RNA auxiliary factor 1-like 4
|
|
chr3_-_191322919
|
0.803
|
NM_001134418
|
LEPREL1
|
leprecan-like 1
|
|
chr17_-_38087366
|
0.802
|
NM_016602
|
CCR10
|
chemokine (C-C motif) receptor 10
|
|
chr1_-_171442937
|
0.800
|
NM_003326
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr14_-_105997732
|
0.796
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr9_+_115246831
|
0.789
|
NM_144488
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr1_+_26516967
|
0.789
|
NM_001803
|
CD52
|
CD52 molecule
|
|
chrX_+_55495262
|
0.789
|
NM_014061
|
MAGEH1
|
melanoma antigen family H, 1
|
|
chr11_+_61316968
|
0.788
|
|
FEN1
|
flap structure-specific endonuclease 1
|
|
chrX_+_78087484
|
0.788
|
NM_014499
NM_198333
|
P2RY10
|
purinergic receptor P2Y, G-protein coupled, 10
|
|
chr11_+_61316928
|
0.787
|
|
FEN1
|
flap structure-specific endonuclease 1
|
|
chr2_-_88946767
|
0.784
|
|
|
|
|
chr2_-_74259308
|
0.784
|
NM_018221
|
MOBKL1B
|
MOB1, Mps One Binder kinase activator-like 1B (yeast)
|
|
chr11_+_61316989
|
0.784
|
|
FEN1
|
flap structure-specific endonuclease 1
|
|
chr22_+_21543671
|
0.780
|
|
IGLV4-3
|
immunoglobulin lambda variable 4-3
|
|
chr20_+_44180355
|
0.777
|
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5
|