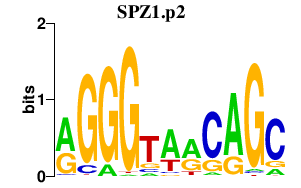
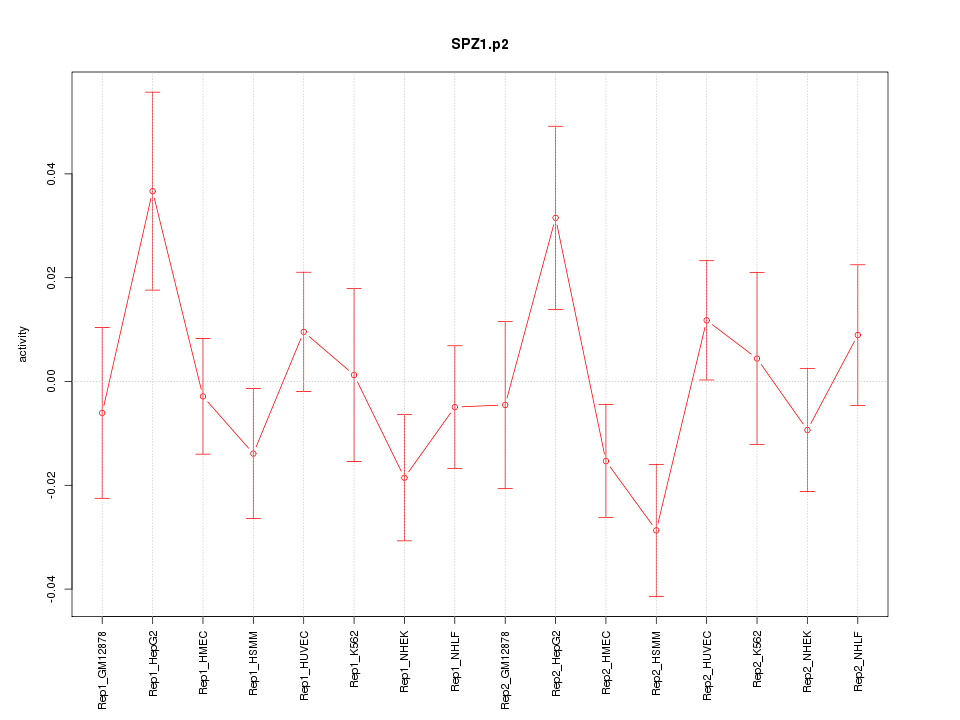
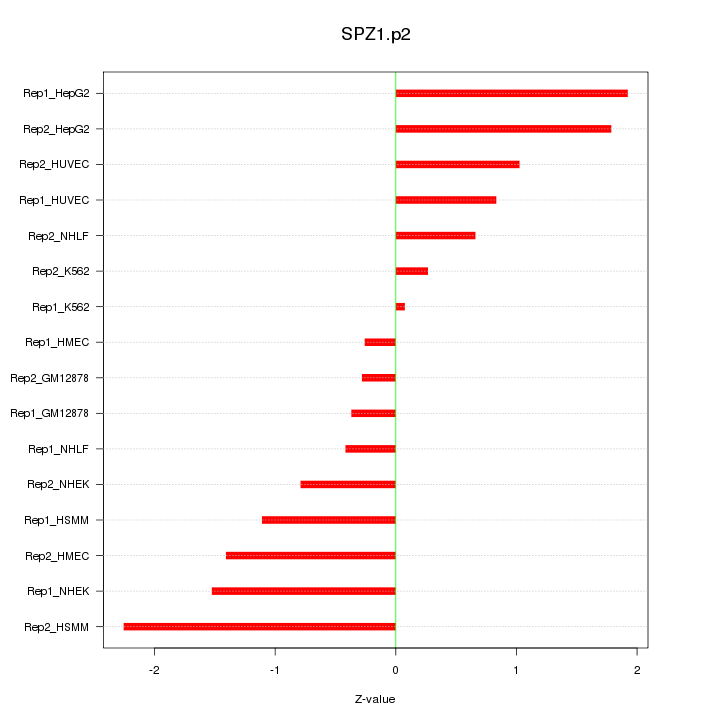
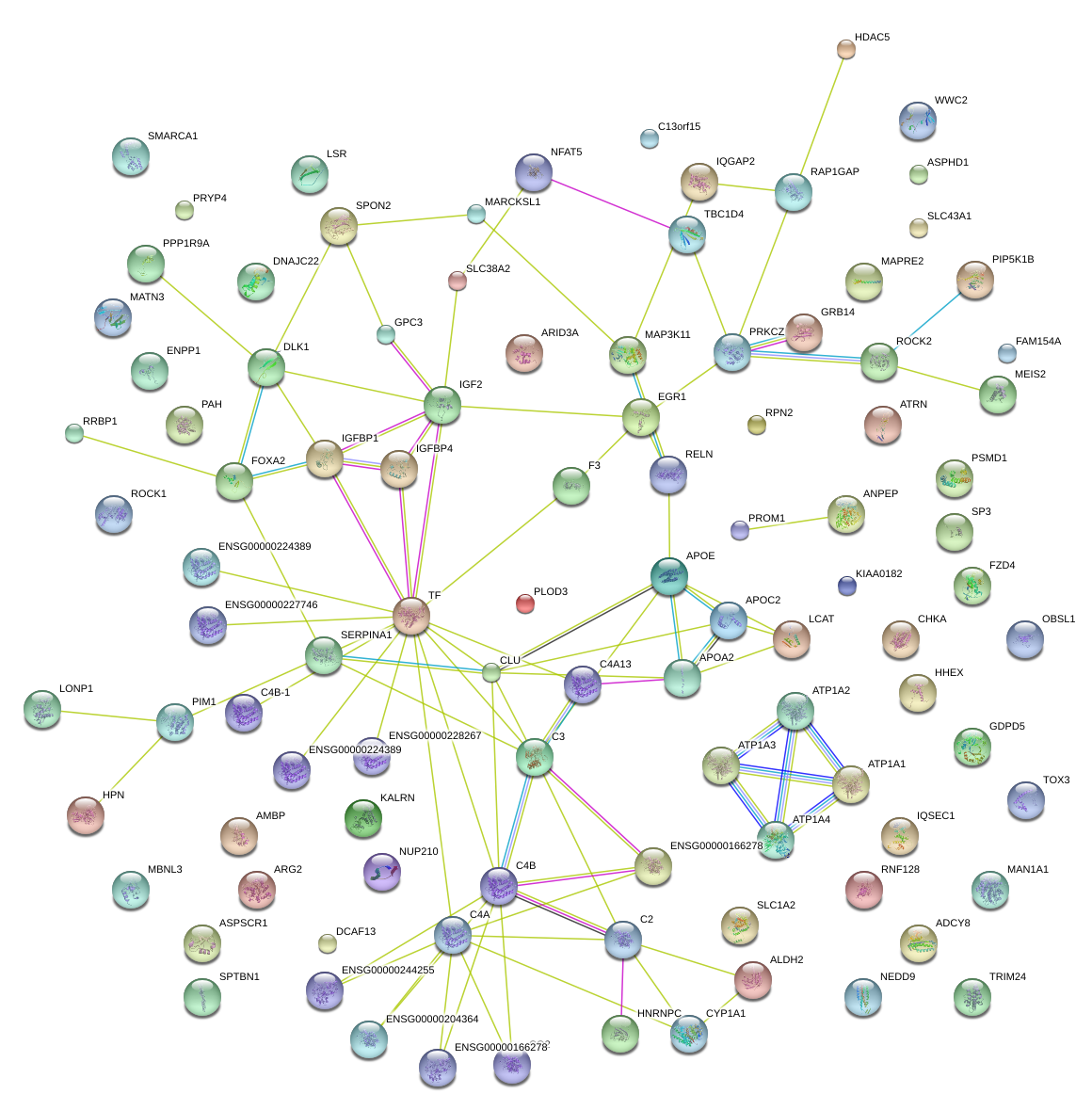
|
chrX_-_132947141
|
2.107
|
|
GPC3
|
glypican 3
|
|
chrX_-_132947290
|
2.057
|
NM_001164617
NM_001164618
NM_001164619
NM_004484
|
GPC3
|
glypican 3
|
|
chr7_-_103417198
|
1.664
|
NM_005045
NM_173054
|
RELN
|
reelin
|
|
chr7_+_45894480
|
1.565
|
NM_000596
|
IGFBP1
|
insulin-like growth factor binding protein 1
|
|
chr6_-_119712596
|
1.550
|
NM_005907
|
MAN1A1
|
mannosidase, alpha, class 1A, member 1
|
|
chr6_+_37245860
|
1.414
|
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr1_-_32574185
|
1.395
|
NM_023009
|
MARCKSL1
|
MARCKS-like 1
|
|
chr1_-_159459999
|
1.371
|
NM_001643
|
APOA2
|
apolipoprotein A-II
|
|
chr13_-_74953873
|
1.247
|
NM_014832
|
TBC1D4
|
TBC1 domain family, member 4
|
|
chr14_+_100270222
|
1.103
|
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chr12_+_110688728
|
1.095
|
NM_000690
|
ALDH2
|
aldehyde dehydrogenase 2 family (mitochondrial)
|
|
chr6_+_32090516
|
1.082
|
NM_007293
NM_001002029
|
C4A
C4B
|
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr12_+_48026955
|
1.043
|
|
DNAJC22
|
DnaJ (Hsp40) homolog, subfamily C, member 22
|
|
chr2_-_20075889
|
1.035
|
|
MATN3
|
matrilin 3
|
|
chr11_-_57039721
|
0.979
|
NM_003627
|
SLC43A1
|
solute carrier family 43, member 1
|
|
chr16_+_29373365
|
0.923
|
NM_001014999
NM_001015000
NM_024044
NM_178044
|
SLX1A-SULT1A3
SLX1A
SLX1B
|
SLX1A-SULT1A3 read-through transcript
SLX1 structure-specific endonuclease subunit homolog A (S. cerevisiae)
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae)
|
|
chr2_-_220144254
|
0.898
|
|
OBSL1
|
obscurin-like 1
|
|
chr2_-_20075929
|
0.892
|
NM_002381
|
MATN3
|
matrilin 3
|
|
chr2_-_174538197
|
0.887
|
|
SP3
|
Sp3 transcription factor
|
|
chrX_-_128485121
|
0.886
|
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chr4_+_184257337
|
0.880
|
NM_024949
|
WWC2
|
WW and C2 domain containing 2
|
|
chr6_-_11490487
|
0.871
|
NM_001142393
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr11_-_35397090
|
0.862
|
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr7_+_137795719
|
0.861
|
|
TRIM24
|
tripartite motif containing 24
|
|
chr19_+_40223249
|
0.860
|
NM_002151
NM_182983
|
HPN
|
hepsin
|
|
chr11_-_2116775
|
0.854
|
NM_000612
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chrX_+_105856531
|
0.810
|
NM_194463
|
RNF128
|
ring finger protein 128
|
|
chr11_-_86343859
|
0.809
|
NM_012193
|
FZD4
|
frizzled homolog 4 (Drosophila)
|
|
chr20_-_22513050
|
0.809
|
NM_021784
|
FOXA2
|
forkhead box A2
|
|
chr7_+_94374862
|
0.804
|
NM_001166160
NM_017650
|
PPP1R9A
|
protein phosphatase 1, regulatory (inhibitor) subunit 9A
|
|
chr16_-_51138306
|
0.777
|
NM_001080430
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr7_-_100647585
|
0.769
|
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3
|
|
chr9_+_70509925
|
0.762
|
NM_003558
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta
|
|
chr20_-_17610704
|
0.739
|
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr7_-_100647439
|
0.723
|
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3
|
|
chr17_-_39556483
|
0.716
|
|
HDAC5
|
histone deacetylase 5
|
|
chr20_-_17610828
|
0.704
|
NM_001042576
NM_004587
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr16_+_29819647
|
0.702
|
NM_181718
|
ASPHD1
|
aspartate beta-hydroxylase domain containing 1
|
|
chr11_-_65138217
|
0.702
|
NM_002419
|
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11
|
|
chr6_+_132170888
|
0.692
|
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chr7_-_100647335
|
0.689
|
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3
|
|
chr13_+_40929525
|
0.683
|
NM_014059
|
C13orf15
|
chromosome 13 open reading frame 15
|
|
chr15_-_72804763
|
0.677
|
NM_000499
|
CYP1A1
|
cytochrome P450, family 1, subfamily A, polypeptide 1
|
|
chr5_+_137829077
|
0.677
|
NM_001964
|
EGR1
|
early growth response 1
|
|
chr18_+_30812205
|
0.671
|
NM_001143827
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr1_-_21868380
|
0.671
|
NM_001145657
NM_002885
|
RAP1GAP
|
RAP1 GTPase activating protein
|
|
chr9_-_115880366
|
0.663
|
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr19_-_6671585
|
0.660
|
NM_000064
|
C3
|
complement component 3
|
|
chr3_+_125786195
|
0.653
|
NM_007064
|
KALRN
|
kalirin, RhoGEF kinase
|
|
chr1_-_159459814
|
0.652
|
|
APOA2
|
apolipoprotein A-II
|
|
chr19_+_876984
|
0.650
|
NM_005224
|
ARID3A
|
AT rich interactive domain 3A (BRIGHT-like)
|
|
chr11_-_67645433
|
0.640
|
NM_001277
NM_212469
|
CHKA
|
choline kinase alpha
|
|
chr15_-_35178888
|
0.635
|
NM_172316
|
MEIS2
|
Meis homeobox 2
|
|
chr16_-_66535457
|
0.626
|
NM_000229
|
LCAT
|
lecithin-cholesterol acyltransferase
|
|
chr6_+_31976754
|
0.622
|
NM_001178063
|
C2
|
complement component 2
|
|
chr7_-_100647561
|
0.613
|
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3
|
|
chr2_-_165186232
|
0.611
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr15_-_20929608
|
0.610
|
|
HERC2P2
|
hect domain and RLD 2 pseudogene 2
|
|
chr4_-_15694407
|
0.607
|
|
PROM1
|
prominin 1
|
|
chr3_+_134947788
|
0.606
|
|
TF
|
transferrin
|
|
chr2_+_54536780
|
0.605
|
NM_003128
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|
|
chr9_-_115880281
|
0.603
|
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr4_-_1156362
|
0.603
|
|
SPON2
|
spondin 2, extracellular matrix protein
|
|
chr8_+_43067817
|
0.594
|
|
|
|
|
chr20_-_22514092
|
0.594
|
NM_153675
|
FOXA2
|
forkhead box A2
|
|
chr14_-_93914634
|
0.592
|
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr2_-_11402150
|
0.590
|
NM_004850
|
ROCK2
|
Rho-associated, coiled-coil containing protein kinase 2
|
|
chr2_-_165186064
|
0.590
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr9_-_115880372
|
0.586
|
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr11_-_2116501
|
0.582
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr13_+_40929679
|
0.579
|
|
C13orf15
|
chromosome 13 open reading frame 15
|
|
chr20_-_22512893
|
0.577
|
|
FOXA2
|
forkhead box A2
|
|
chr17_+_35853177
|
0.569
|
NM_001552
|
IGFBP4
|
insulin-like growth factor binding protein 4
|
|
chr3_+_134947573
|
0.561
|
NM_001063
|
TF
|
transferrin
|
|
chr15_-_88150823
|
0.560
|
|
ANPEP
|
alanyl (membrane) aminopeptidase
|
|
chr13_+_40929690
|
0.560
|
|
C13orf15
|
chromosome 13 open reading frame 15
|
|
chr11_-_74914514
|
0.556
|
|
GDPD5
|
glycerophosphodiester phosphodiesterase domain containing 5
|
|
chr1_+_116717461
|
0.551
|
|
ATP1A1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide
|
|
chr6_+_32057779
|
0.547
|
NM_007293
NM_001002029
|
C4A
C4B
|
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr12_-_45052830
|
0.547
|
NM_018976
|
SLC38A2
|
solute carrier family 38, member 2
|
|
chrX_-_131451675
|
0.545
|
NM_001170704
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr14_+_67156386
|
0.545
|
|
ARG2
|
arginase, type II
|
|
chr19_+_50100843
|
0.541
|
NM_000041
|
APOE
|
apolipoprotein E
|
|
chr16_+_68157611
|
0.535
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr9_-_115880425
|
0.534
|
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr12_-_101835026
|
0.528
|
|
PAH
|
phenylalanine hydroxylase
|
|
chr6_-_167289659
|
0.526
|
|
|
|
|
chr10_+_94439658
|
0.520
|
NM_002729
|
HHEX
|
hematopoietically expressed homeobox
|
|
chr12_-_101835510
|
0.519
|
NM_000277
|
PAH
|
phenylalanine hydroxylase
|
|
chr3_-_13436802
|
0.517
|
NM_024923
|
NUP210
|
nucleoporin 210kDa
|
|
chr8_-_27528215
|
0.512
|
NM_001831
|
CLU
|
clusterin
|
|
chr4_-_15694691
|
0.512
|
NM_001145847
NM_001145848
|
PROM1
|
prominin 1
|
|
chr17_+_77528699
|
0.512
|
NM_024083
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1
|
|
chr1_+_1994943
|
0.508
|
NM_001033581
|
PRKCZ
|
protein kinase C, zeta
|
|
chr5_+_75734759
|
0.505
|
NM_006633
|
IQGAP2
|
IQ motif containing GTPase activating protein 2
|
|
chr19_+_50144267
|
0.502
|
|
APOC2
|
apolipoprotein C-II
|
|
chrX_-_128485062
|
0.500
|
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chr19_+_40431678
|
0.497
|
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr9_-_115880494
|
0.496
|
NM_001633
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr3_-_13089547
|
0.488
|
NM_001134382
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr16_+_84202515
|
0.486
|
NM_001134473
|
KIAA0182
|
KIAA0182
|
|
chr20_+_35241161
|
0.476
|
|
RPN2
|
ribophorin II
|
|
chr20_+_3399615
|
0.474
|
NM_139321
NM_139322
|
ATRN
|
attractin
|
|
chr7_-_30600868
|
0.474
|
|
LOC285949
|
hypothetical protein LOC285949
|
|
chr1_-_11909042
|
0.473
|
NM_138346
|
KIAA2013
|
KIAA2013
|
|
chr4_-_1156378
|
0.468
|
|
SPON2
|
spondin 2, extracellular matrix protein
|
|
chr4_-_1156458
|
0.464
|
|
SPON2
|
spondin 2, extracellular matrix protein
|
|
chr2_-_101134146
|
0.464
|
NM_001102426
|
TBC1D8
|
TBC1 domain family, member 8 (with GRAM domain)
|
|
chr12_-_101835183
|
0.462
|
|
PAH
|
phenylalanine hydroxylase
|
|
chr10_+_114700775
|
0.458
|
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr18_-_50777634
|
0.457
|
NM_025214
|
CCDC68
|
coiled-coil domain containing 68
|
|
chr2_-_165186603
|
0.456
|
NM_004490
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr15_-_20666684
|
0.454
|
|
LOC283683
|
hypothetical LOC283683
|
|
chr1_+_39229474
|
0.450
|
NM_001136275
NM_024595
|
AKIRIN1
|
akirin 1
|
|
chr11_-_70185559
|
0.443
|
NM_133266
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr3_-_187025501
|
0.442
|
NM_001007225
NM_006548
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr2_-_174537354
|
0.442
|
|
SP3
|
Sp3 transcription factor
|
|
chr3_-_50335113
|
0.442
|
NM_003773
NM_033158
|
HYAL2
|
hyaluronoglucosaminidase 2
|
|
chr4_-_88531353
|
0.438
|
NM_016245
|
HSD17B11
|
hydroxysteroid (17-beta) dehydrogenase 11
|
|
chr4_-_88531245
|
0.437
|
|
HSD17B11
|
hydroxysteroid (17-beta) dehydrogenase 11
|
|
chr8_-_17599438
|
0.433
|
NM_020749
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr6_-_52549673
|
0.430
|
|
TRAM2
|
translocation associated membrane protein 2
|
|
chr6_-_52549774
|
0.425
|
NM_012288
|
TRAM2
|
translocation associated membrane protein 2
|
|
chr1_-_92124201
|
0.423
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr2_+_159021653
|
0.421
|
|
PKP4
|
plakophilin 4
|
|
chr1_-_56817802
|
0.419
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr5_-_175896829
|
0.416
|
NM_014901
|
RNF44
|
ring finger protein 44
|
|
chr12_-_24606616
|
0.415
|
NM_152989
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr2_-_220144427
|
0.415
|
|
OBSL1
|
obscurin-like 1
|
|
chr7_-_100647711
|
0.414
|
NM_001084
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3
|
|
chr19_-_45947667
|
0.412
|
NM_198476
|
C19orf54
|
chromosome 19 open reading frame 54
|
|
chr20_+_35241109
|
0.410
|
|
RPN2
|
ribophorin II
|
|
chr14_+_23937766
|
0.408
|
NM_025081
|
NYNRIN
|
NYN domain and retroviral integrase containing
|
|
chr5_+_92946348
|
0.407
|
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr7_+_94135384
|
0.407
|
|
PEG10
|
paternally expressed 10
|
|
chr5_+_150380325
|
0.406
|
|
GPX3
|
glutathione peroxidase 3 (plasma)
|
|
chr13_+_34414390
|
0.404
|
NM_015678
|
NBEA
|
neurobeachin
|
|
chr1_-_114855281
|
0.403
|
NM_015906
NM_033020
|
TRIM33
|
tripartite motif containing 33
|
|
chr4_-_22126437
|
0.396
|
NM_145290
|
GPR125
|
G protein-coupled receptor 125
|
|
chr1_-_114855200
|
0.395
|
|
TRIM33
|
tripartite motif containing 33
|
|
chr8_-_27528105
|
0.394
|
|
CLU
|
clusterin
|
|
chr7_-_87180499
|
0.394
|
NM_000927
|
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1
|
|
chr20_-_47238082
|
0.390
|
|
STAU1
|
staufen, RNA binding protein, homolog 1 (Drosophila)
|
|
chr5_-_141684759
|
0.389
|
NM_001127496
NM_030964
|
SPRY4
|
sprouty homolog 4 (Drosophila)
|
|
chr4_-_74343320
|
0.387
|
NM_032217
NM_198889
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr19_+_50041387
|
0.386
|
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr18_-_50004458
|
0.385
|
|
MBD2
|
methyl-CpG binding domain protein 2
|
|
chr11_-_74914066
|
0.385
|
NM_030792
|
GDPD5
|
glycerophosphodiester phosphodiesterase domain containing 5
|
|
chr12_+_119456538
|
0.384
|
|
RNF10
|
ring finger protein 10
|
|
chr7_-_99411573
|
0.383
|
NM_001185
|
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding
|
|
chr16_+_29597829
|
0.382
|
NM_014298
|
QPRT
|
quinolinate phosphoribosyltransferase
|
|
chr19_+_50041400
|
0.381
|
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr4_-_74343169
|
0.381
|
|
|
|
|
chr16_-_22293097
|
0.379
|
NM_001802
|
CDR2
|
cerebellar degeneration-related protein 2, 62kDa
|
|
chr17_-_39556535
|
0.378
|
NM_001015053
NM_005474
|
HDAC5
|
histone deacetylase 5
|
|
chr4_+_74493218
|
0.377
|
|
ALB
|
albumin
|
|
chr12_-_12394402
|
0.377
|
NM_018050
|
MANSC1
|
MANSC domain containing 1
|
|
chr16_+_29597965
|
0.375
|
|
QPRT
|
quinolinate phosphoribosyltransferase
|
|
chr12_+_110935618
|
0.374
|
|
ERP29
|
endoplasmic reticulum protein 29
|
|
chr12_-_45052286
|
0.369
|
|
SLC38A2
|
solute carrier family 38, member 2
|
|
chr3_-_95175282
|
0.369
|
|
PROS1
|
protein S (alpha)
|
|
chr6_+_43151922
|
0.369
|
NM_002821
NM_152880
NM_152881
NM_152882
|
PTK7
|
PTK7 protein tyrosine kinase 7
|
|
chr12_+_110935644
|
0.368
|
|
ERP29
|
endoplasmic reticulum protein 29
|
|
chr17_-_71861292
|
0.368
|
|
PRPSAP1
|
phosphoribosyl pyrophosphate synthetase-associated protein 1
|
|
chr11_-_2116360
|
0.363
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr17_+_69939806
|
0.362
|
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C
|
|
chr1_-_180627934
|
0.361
|
NM_001033044
NM_002065
|
GLUL
|
glutamate-ammonia ligase
|
|
chr4_+_74493331
|
0.359
|
|
ALB
|
albumin
|
|
chr14_-_20975241
|
0.356
|
NM_020920
|
CHD8
|
chromodomain helicase DNA binding protein 8
|
|
chr6_-_47385470
|
0.354
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr1_-_180627023
|
0.354
|
NM_001033056
|
GLUL
|
glutamate-ammonia ligase
|
|
chr20_-_32877029
|
0.351
|
NM_014071
|
NCOA6
|
nuclear receptor coactivator 6
|
|
chr10_+_114125927
|
0.349
|
NM_016234
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr2_-_175741096
|
0.348
|
NM_001880
|
ATF2
|
activating transcription factor 2
|
|
chr13_+_30089829
|
0.347
|
NM_005800
|
USPL1
|
ubiquitin specific peptidase like 1
|
|
chr1_-_203867402
|
0.347
|
|
ELK4
|
ELK4, ETS-domain protein (SRF accessory protein 1)
|
|
chrX_+_152606945
|
0.346
|
NM_001142805
NM_005629
|
SLC6A8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8
|
|
chr11_-_57038891
|
0.346
|
NM_001198810
|
SLC43A1
|
solute carrier family 43, member 1
|
|
chr7_+_94130752
|
0.346
|
|
PEG10
|
paternally expressed 10
|
|
chr20_+_35240844
|
0.345
|
NM_001135771
NM_002951
|
RPN2
|
ribophorin II
|
|
chr6_-_47385205
|
0.345
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr10_-_28861465
|
0.343
|
|
LOC220906
|
hypothetical LOC220906
|
|
chr11_-_67645246
|
0.339
|
|
CHKA
|
choline kinase alpha
|
|
chr20_+_35240936
|
0.338
|
|
RPN2
|
ribophorin II
|
|
chr6_-_47385313
|
0.334
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr12_+_110935534
|
0.334
|
NM_001034025
NM_006817
|
ERP29
|
endoplasmic reticulum protein 29
|
|
chr12_+_970613
|
0.333
|
NM_178039
NM_178040
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1
|
|
chr4_-_141568210
|
0.329
|
NM_001130675
NM_004362
|
CLGN
|
calmegin
|
|
chr19_+_50041232
|
0.328
|
NM_001042724
NM_002856
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr14_-_89155078
|
0.328
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chr17_-_50854054
|
0.326
|
NM_012329
|
MMD
|
monocyte to macrophage differentiation-associated
|
|
chr11_-_2116048
|
0.325
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr5_+_154115046
|
0.324
|
|
LARP1
|
La ribonucleoprotein domain family, member 1
|
|
chr1_+_26896519
|
0.323
|
|
ARID1A
|
AT rich interactive domain 1A (SWI-like)
|
|
chr3_+_50281309
|
0.322
|
|
SEMA3B
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B
|
|
chr16_+_28741861
|
0.322
|
|
ATXN2L
|
ataxin 2-like
|
|
chr11_-_76059435
|
0.321
|
NM_001128922
|
LRRC32
|
leucine rich repeat containing 32
|
|
chr16_+_29728932
|
0.320
|
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor)
|
|
chr5_-_172689111
|
0.320
|
NM_003714
|
STC2
|
stanniocalcin 2
|
|
chr17_+_38256710
|
0.319
|
NM_003734
|
AOC3
|
amine oxidase, copper containing 3 (vascular adhesion protein 1)
|