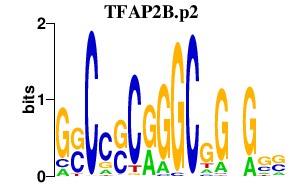
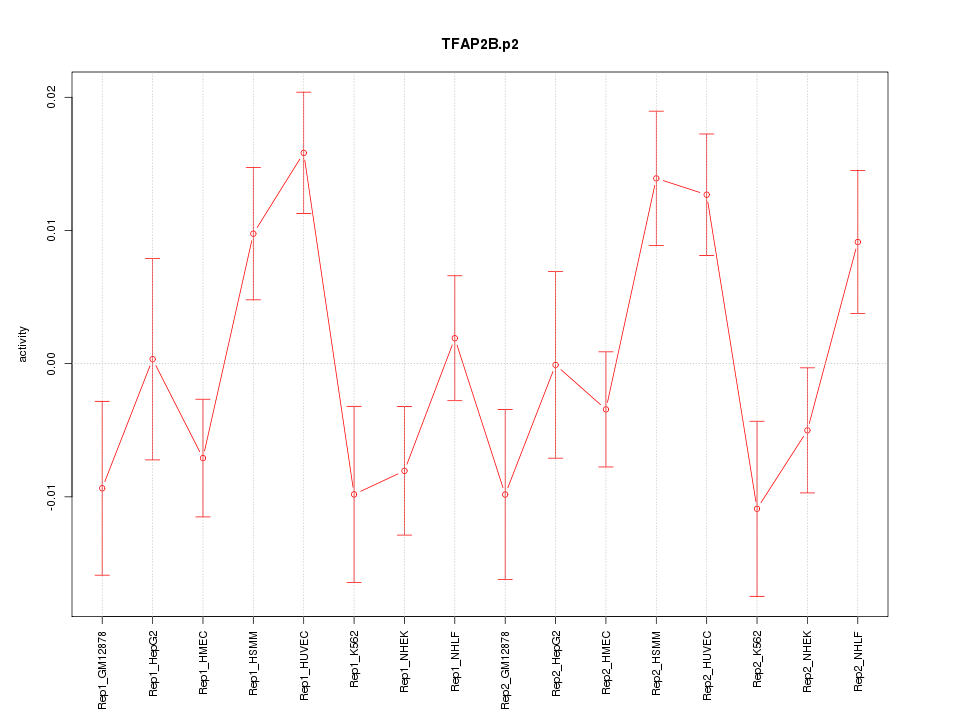
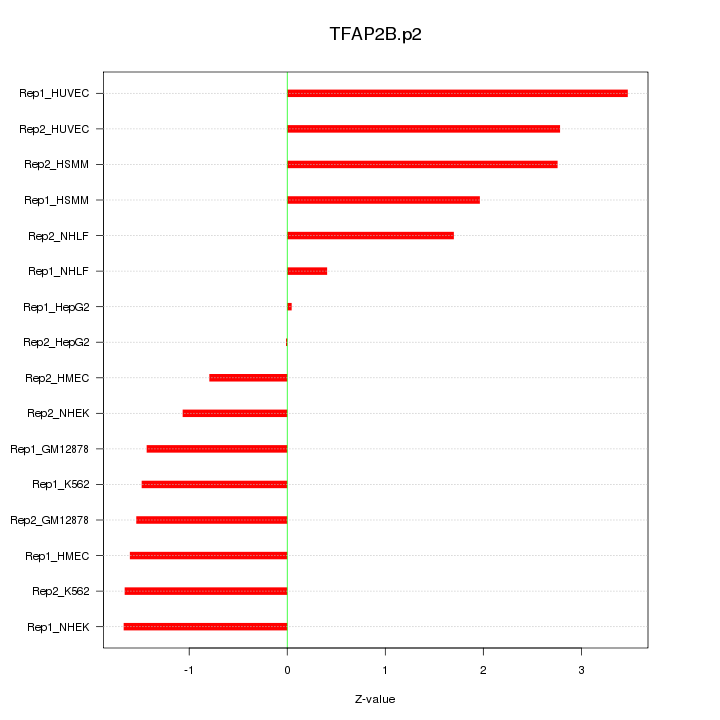
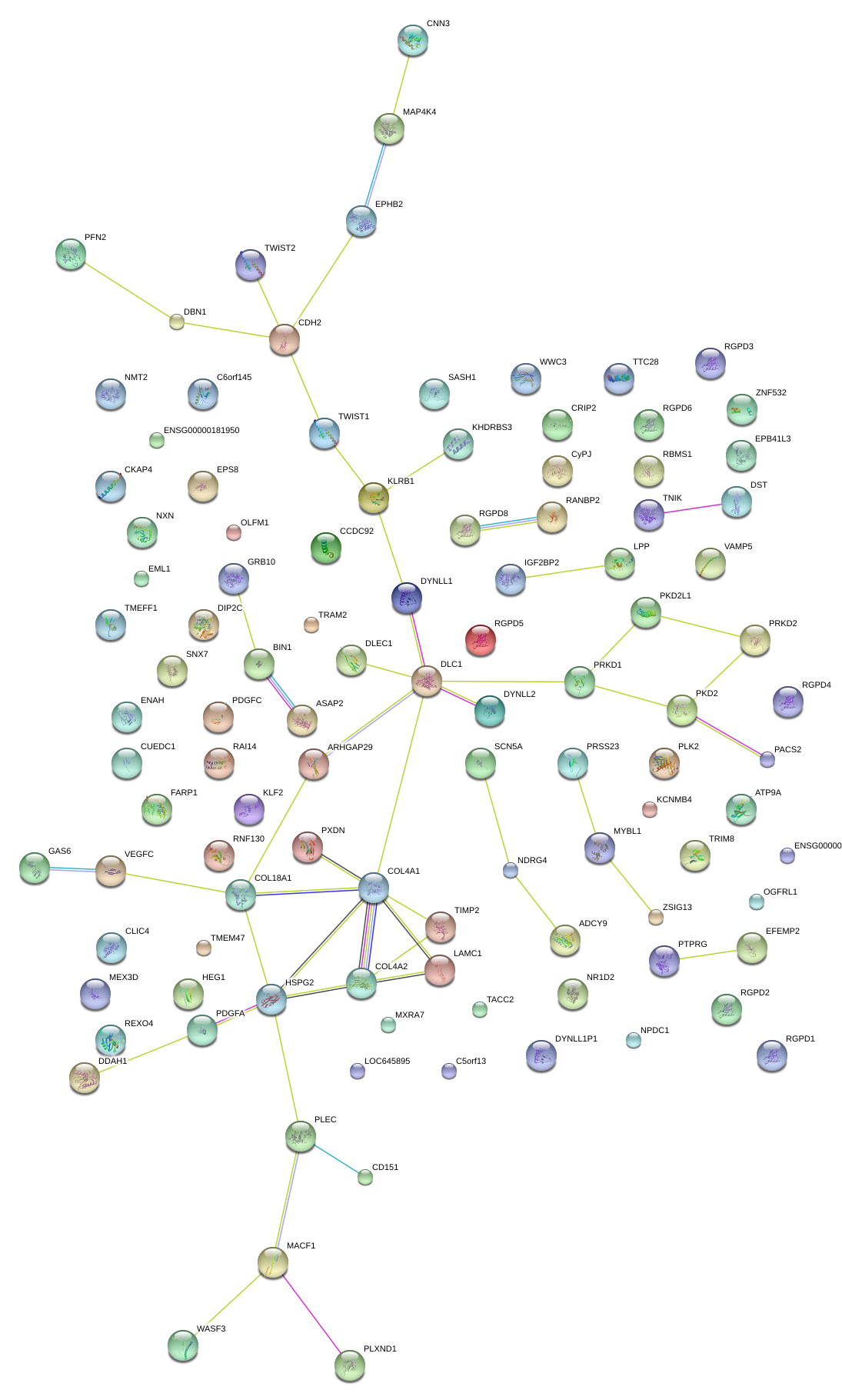
|
chr17_-_74432800
|
3.952
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr2_-_160972606
|
3.923
|
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr17_-_74433048
|
3.222
|
NM_003255
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr3_-_151171190
|
2.837
|
NM_002628
NM_053024
|
PFN2
|
profilin 2
|
|
chr3_-_130807865
|
2.787
|
NM_015103
|
PLXND1
|
plexin D1
|
|
chr14_+_105012106
|
2.724
|
NM_001312
|
CRIP2
|
cysteine-rich protein 2
|
|
chr17_-_74432671
|
2.708
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr18_-_5533967
|
2.666
|
NM_012307
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3
|
|
chr1_-_95165037
|
2.562
|
|
CNN3
|
calponin 3, acidic
|
|
chr11_-_65396775
|
2.468
|
NM_016938
|
EFEMP2
|
EGF containing fibulin-like extracellular matrix protein 2
|
|
chr18_-_24011349
|
2.360
|
NM_001792
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr20_-_49818268
|
2.331
|
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr1_-_95165089
|
2.265
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr12_-_105165805
|
2.133
|
NM_006825
|
CKAP4
|
cytoskeleton-associated protein 4
|
|
chr8_+_136538739
|
2.088
|
NM_006558
|
KHDRBS3
|
KH domain containing, RNA binding, signal transduction associated 3
|
|
chr1_+_24944554
|
2.059
|
|
CLIC4
|
chloride intracellular channel 4
|
|
chr1_-_85703321
|
2.054
|
NM_012137
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chrX_+_9943793
|
2.050
|
NM_015691
|
WWC3
|
WWC family member 3
|
|
chr14_-_29466562
|
1.994
|
NM_002742
|
PRKD1
|
protein kinase D1
|
|
chr20_-_49818306
|
1.991
|
NM_006045
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr6_+_72055229
|
1.960
|
|
OGFRL1
|
opioid growth factor receptor-like 1
|
|
chr1_+_181258952
|
1.932
|
NM_002293
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2)
|
|
chr21_+_45649429
|
1.863
|
NM_130445
|
COL18A1
|
collagen, type XVIII, alpha 1
|
|
chr12_+_69046241
|
1.835
|
NM_014505
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4
|
|
chr7_-_50828159
|
1.814
|
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chr4_+_89147821
|
1.796
|
NM_000297
|
PKD2
|
polycystic kidney disease 2 (autosomal dominant)
|
|
chr3_-_151171576
|
1.760
|
|
PFN2
|
profilin 2
|
|
chr5_-_111121150
|
1.760
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr1_-_85703198
|
1.759
|
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr1_+_181259317
|
1.741
|
|
|
|
|
chr6_+_72055197
|
1.735
|
NM_024576
|
OGFRL1
|
opioid growth factor receptor-like 1
|
|
chr10_+_104394185
|
1.734
|
NM_030912
|
TRIM8
|
tripartite motif containing 8
|
|
chr7_-_50828608
|
1.722
|
NM_001001555
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chr3_-_38666122
|
1.712
|
NM_000335
NM_001099404
NM_001099405
NM_198056
NM_001160160
NM_001160161
|
SCN5A
|
sodium channel, voltage-gated, type V, alpha subunit
|
|
chr3_-_126257425
|
1.711
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr6_+_72055235
|
1.694
|
|
OGFRL1
|
opioid growth factor receptor-like 1
|
|
chr11_+_86189138
|
1.628
|
NM_007173
|
PRSS23
|
protease, serine, 23
|
|
chr4_-_177950658
|
1.614
|
NM_005429
|
VEGFC
|
vascular endothelial growth factor C
|
|
chr2_-_1727247
|
1.600
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr5_+_34692239
|
1.579
|
|
RAI14
|
retinoic acid induced 14
|
|
chr2_-_161058120
|
1.574
|
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr5_-_176833259
|
1.567
|
NM_004395
|
DBN1
|
drebrin 1
|
|
chr9_+_137119339
|
1.566
|
|
OLFM1
|
olfactomedin 1
|
|
chr13_+_97593469
|
1.554
|
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr6_-_52549774
|
1.553
|
NM_012288
|
TRAM2
|
translocation associated membrane protein 2
|
|
chr2_+_9264360
|
1.543
|
|
ASAP2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2
|
|
chr6_-_52549673
|
1.524
|
|
TRAM2
|
translocation associated membrane protein 2
|
|
chr13_+_97593411
|
1.519
|
NM_001001715
NM_005766
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr5_-_176833197
|
1.473
|
|
DBN1
|
drebrin 1
|
|
chr3_-_187025501
|
1.472
|
NM_001007225
NM_006548
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr19_+_16296632
|
1.470
|
NM_016270
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr5_+_34692352
|
1.466
|
NM_001145520
|
RAI14
|
retinoic acid induced 14
|
|
chr1_+_39319675
|
1.457
|
NM_012090
|
MACF1
|
microtubule-actin crosslinking factor 1
|
|
chr13_-_109757442
|
1.434
|
NM_001845
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr8_-_67687980
|
1.387
|
NM_001080416
NM_001144755
|
MYBL1
LOC645895
|
v-myb myeloblastosis viral oncogene homolog (avian)-like 1
hypothetical LOC645895
|
|
chr9_+_102275455
|
1.385
|
|
TMEFF1
|
transmembrane protein with EGF-like and two follistatin-like domains 1
|
|
chr1_+_24944346
|
1.354
|
NM_013943
|
CLIC4
|
chloride intracellular channel 4
|
|
chr13_+_97593711
|
1.343
|
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr21_+_45649719
|
1.342
|
|
|
|
|
chr13_+_109757593
|
1.332
|
NM_001846
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr1_+_24944493
|
1.328
|
|
CLIC4
|
chloride intracellular channel 4
|
|
chr3_+_189354167
|
1.323
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr2_-_127580975
|
1.300
|
NM_004305
NM_139343
NM_139344
NM_139345
NM_139346
NM_139347
NM_139348
NM_139349
NM_139350
NM_139351
|
BIN1
|
bridging integrator 1
|
|
chr22_-_27405708
|
1.277
|
NM_001145418
|
TTC28
|
tetratricopeptide repeat domain 28
|
|
chr6_+_148705646
|
1.275
|
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr1_+_98899734
|
1.261
|
NM_015976
NM_152238
|
SNX7
|
sorting nexin 7
|
|
chr9_+_102275310
|
1.261
|
NM_003692
|
TMEFF1
|
transmembrane protein with EGF-like and two follistatin-like domains 1
|
|
chr1_-_94475504
|
1.255
|
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chrX_-_34585287
|
1.248
|
NM_031442
|
TMEM47
|
transmembrane protein 47
|
|
chr3_+_61522219
|
1.234
|
NM_002841
|
PTPRG
|
protein tyrosine phosphatase, receptor type, G
|
|
chr13_+_113546943
|
1.226
|
|
GAS6
|
growth arrest-specific 6
|
|
chr2_-_1727324
|
1.221
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr1_-_94475840
|
1.207
|
NM_004815
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr5_-_111121691
|
1.176
|
NM_001142477
NM_001142476
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr10_-_725522
|
1.167
|
NM_014974
|
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila)
|
|
chr2_+_9264327
|
1.164
|
NM_001135191
NM_003887
|
ASAP2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2
|
|
chr19_-_1518875
|
1.164
|
NM_001174118
NM_203304
|
MEX3D
|
mex-3 homolog D (C. elegans)
|
|
chr8_+_136539290
|
1.162
|
|
KHDRBS3
|
KH domain containing, RNA binding, signal transduction associated 3
|
|
chr10_+_123913094
|
1.161
|
NM_006997
NM_206860
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr10_+_123912930
|
1.156
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr14_+_104852225
|
1.153
|
|
PACS2
|
phosphofurin acidic cluster sorting protein 2
|
|
chr1_+_22909916
|
1.149
|
NM_004442
NM_017449
|
EPHB2
|
EPH receptor B2
|
|
chr2_-_110769670
|
1.134
|
NM_001164463
NM_005054
|
RGPD8
RGPD5
|
RANBP2-like and GRIP domain containing 8
RANBP2-like and GRIP domain containing 5
|
|
chr17_-_72218661
|
1.133
|
|
MXRA7
|
matrix-remodelling associated 7
|
|
chr2_-_1727292
|
1.126
|
NM_012293
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr10_-_15250616
|
1.126
|
NM_004808
|
NMT2
|
N-myristoyltransferase 2
|
|
chr17_-_53387493
|
1.118
|
|
CUEDC1
|
CUE domain containing 1
|
|
chr12_-_123023312
|
1.117
|
|
CCDC92
|
coiled-coil domain containing 92
|
|
chr18_+_54681657
|
1.102
|
|
ZNF532
|
zinc finger protein 532
|
|
chr14_+_99329198
|
1.099
|
NM_001008707
NM_004434
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr9_-_139060416
|
1.089
|
NM_015392
|
NPDC1
|
neural proliferation, differentiation and control, 1
|
|
chr1_-_22136263
|
1.087
|
NM_005529
|
HSPG2
|
heparan sulfate proteoglycan 2
|
|
chr2_+_85665041
|
1.083
|
NM_006634
|
VAMP5
|
vesicle-associated membrane protein 5 (myobrevin)
|
|
chr2_+_101680594
|
1.073
|
NM_004834
NM_145686
NM_145687
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr5_-_111120929
|
1.064
|
NM_001142482
NM_001142481
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr9_-_139060285
|
1.063
|
|
NPDC1
|
neural proliferation, differentiation and control, 1
|
|
chr5_-_57791544
|
1.060
|
NM_006622
|
PLK2
|
polo-like kinase 2
|
|
chr5_+_34692124
|
1.058
|
NM_001145522
NM_015577
|
RAI14
|
retinoic acid induced 14
|
|
chr4_-_158111995
|
1.049
|
NM_016205
|
PDGFC
|
platelet derived growth factor C
|
|
chr7_-_19123765
|
1.044
|
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr12_-_15833617
|
1.038
|
NM_004447
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr11_+_822965
|
1.038
|
|
CD151
|
CD151 molecule (Raph blood group)
|
|
chr9_-_139060377
|
1.034
|
|
NPDC1
|
neural proliferation, differentiation and control, 1
|
|
chr1_-_223907283
|
1.024
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr12_-_15833383
|
1.012
|
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr16_+_57055494
|
1.006
|
NM_001130487
|
NDRG4
|
NDRG family member 4
|
|
chr7_-_19123787
|
0.996
|
NM_000474
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr6_-_3697244
|
0.991
|
NM_183373
|
C6orf145
|
chromosome 6 open reading frame 145
|
|
chr3_+_23961739
|
0.986
|
NM_005126
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr13_+_26029879
|
0.985
|
|
WASF3
|
WAS protein family, member 3
|
|
chr16_-_4106186
|
0.978
|
NM_001116
|
ADCY9
|
adenylate cyclase 9
|
|
chr5_-_179431204
|
0.976
|
|
RNF130
|
ring finger protein 130
|
|
chr17_-_829655
|
0.974
|
NM_022463
|
NXN
|
nucleoredoxin
|
|
chr6_+_148705817
|
0.963
|
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr13_+_113546912
|
0.963
|
|
GAS6
|
growth arrest-specific 6
|
|
chr7_-_525556
|
0.953
|
|
PDGFA
|
platelet-derived growth factor alpha polypeptide
|
|
chr8_-_13035102
|
0.953
|
NM_006094
|
DLC1
|
deleted in liver cancer 1
|
|
chr12_-_104153920
|
0.951
|
NM_018171
|
APPL2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2
|
|
chr4_-_7991992
|
0.951
|
NM_001134647
NM_198595
|
AFAP1
|
actin filament associated protein 1
|
|
chr2_-_39517795
|
0.949
|
|
MAP4K3
|
mitogen-activated protein kinase kinase kinase kinase 3
|
|
chr9_+_136673464
|
0.948
|
NM_000093
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr16_-_22293097
|
0.946
|
NM_001802
|
CDR2
|
cerebellar degeneration-related protein 2, 62kDa
|
|
chr11_+_12355600
|
0.945
|
NM_018222
|
PARVA
|
parvin, alpha
|
|
chr12_-_122322117
|
0.944
|
NM_004642
|
CDK2AP1
|
cyclin-dependent kinase 2 associated protein 1
|
|
chr5_-_179431491
|
0.937
|
|
RNF130
|
ring finger protein 130
|
|
chr17_-_60088773
|
0.937
|
NM_022739
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2
|
|
chr6_+_148705421
|
0.936
|
NM_015278
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr11_+_822909
|
0.934
|
NM_001039490
NM_004357
NM_139029
NM_139030
|
CD151
|
CD151 molecule (Raph blood group)
|
|
chr22_+_36472201
|
0.926
|
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr4_+_41057507
|
0.925
|
NM_001112717
NM_001112718
NM_014988
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr22_+_36472262
|
0.925
|
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr13_-_39075304
|
0.923
|
|
LHFP
|
lipoma HMGIC fusion partner
|
|
chr15_+_37660568
|
0.923
|
NM_003246
|
THBS1
|
thrombospondin 1
|
|
chr14_+_104851948
|
0.922
|
NM_001100913
NM_015197
|
PACS2
|
phosphofurin acidic cluster sorting protein 2
|
|
chr11_-_2863536
|
0.920
|
NM_000076
NM_001122630
NM_001122631
|
CDKN1C
|
cyclin-dependent kinase inhibitor 1C (p57, Kip2)
|
|
chr21_+_41461962
|
0.917
|
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chr7_-_150576660
|
0.911
|
NM_001003801
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3
|
|
chr7_-_2850175
|
0.909
|
NM_007353
|
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12
|
|
chr1_-_43691932
|
0.900
|
|
HYI
|
hydroxypyruvate isomerase (putative)
|
|
chr11_-_86343859
|
0.899
|
NM_012193
|
FZD4
|
frizzled homolog 4 (Drosophila)
|
|
chr7_+_98084531
|
0.895
|
NM_002523
|
NPTX2
|
neuronal pentraxin II
|
|
chr19_+_16296734
|
0.895
|
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr17_-_5345167
|
0.889
|
|
LOC728392
|
hypothetical protein LOC728392
|
|
chr2_+_187162969
|
0.884
|
NM_001145000
NM_002210
|
ITGAV
|
integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51)
|
|
chr8_+_55533047
|
0.883
|
NM_022454
|
SOX17
|
SRY (sex determining region Y)-box 17
|
|
chr3_-_129024665
|
0.876
|
NM_001003794
|
MGLL
|
monoglyceride lipase
|
|
chr19_+_40213431
|
0.871
|
NM_001037
NM_199037
|
SCN1B
|
sodium channel, voltage-gated, type I, beta
|
|
chr19_+_3045528
|
0.867
|
|
GNA11
|
guanine nucleotide binding protein (G protein), alpha 11 (Gq class)
|
|
chr12_-_108802561
|
0.855
|
NM_016433
|
GLTP
|
glycolipid transfer protein
|
|
chr13_+_113546897
|
0.854
|
|
GAS6
|
growth arrest-specific 6
|
|
chr9_+_36026900
|
0.853
|
NM_021111
|
RECK
|
reversion-inducing-cysteine-rich protein with kazal motifs
|
|
chr9_+_127549437
|
0.853
|
NM_006195
|
PBX3
|
pre-B-cell leukemia homeobox 3
|
|
chr4_-_81213207
|
0.848
|
|
|
|
|
chr19_-_18578577
|
0.846
|
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr12_+_51777684
|
0.846
|
NM_002178
|
IGFBP6
|
insulin-like growth factor binding protein 6
|
|
chr8_+_38763911
|
0.837
|
|
TACC1
|
transforming, acidic coiled-coil containing protein 1
|
|
chr11_+_823007
|
0.834
|
|
CD151
|
CD151 molecule (Raph blood group)
|
|
chr5_-_39460687
|
0.831
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr16_+_57055129
|
0.830
|
|
NDRG4
|
NDRG family member 4
|
|
chr12_+_110328126
|
0.830
|
NM_005475
|
SH2B3
|
SH2B adaptor protein 3
|
|
chr1_-_65204414
|
0.827
|
NM_002227
|
JAK1
|
Janus kinase 1
|
|
chr16_+_57055049
|
0.827
|
NM_020465
|
NDRG4
|
NDRG family member 4
|
|
chr11_+_12652427
|
0.827
|
NM_021961
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr4_-_9727456
|
0.820
|
|
WDR1
|
WD repeat domain 1
|
|
chr5_+_102229425
|
0.815
|
NM_000919
NM_001177306
NM_138766
NM_138821
NM_138822
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr3_-_126088530
|
0.815
|
|
ITGB5
|
integrin, beta 5
|
|
chr5_-_176832476
|
0.813
|
NM_080881
|
DBN1
|
drebrin 1
|
|
chr15_+_61121806
|
0.808
|
NM_000366
NM_001018004
NM_001018005
NM_001018006
NM_001018007
NM_001018020
|
TPM1
|
tropomyosin 1 (alpha)
|
|
chr20_-_23566325
|
0.806
|
NM_000099
|
CST3
|
cystatin C
|
|
chr2_+_46377952
|
0.805
|
NM_001430
|
EPAS1
|
endothelial PAS domain protein 1
|
|
chr17_-_75808680
|
0.803
|
NM_000199
|
SGSH
|
N-sulfoglucosamine sulfohydrolase
|
|
chr12_+_51777715
|
0.803
|
|
IGFBP6
|
insulin-like growth factor binding protein 6
|
|
chr19_+_38377329
|
0.802
|
NM_002333
|
LRP3
|
low density lipoprotein receptor-related protein 3
|
|
chrX_-_15782803
|
0.802
|
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr12_-_78608154
|
0.801
|
|
PAWR
|
PRKC, apoptosis, WT1, regulator
|
|
chr8_-_11763023
|
0.797
|
|
CTSB
|
cathepsin B
|
|
chr9_-_87904280
|
0.796
|
|
GOLM1
|
golgi membrane protein 1
|
|
chr8_-_18915443
|
0.795
|
NM_015310
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr8_+_30361485
|
0.793
|
NM_001008710
NM_001008711
NM_001008712
NM_006867
|
RBPMS
|
RNA binding protein with multiple splicing
|
|
chr9_-_87904215
|
0.793
|
|
GOLM1
|
golgi membrane protein 1
|
|
chr9_-_87904306
|
0.792
|
NM_016548
|
GOLM1
|
golgi membrane protein 1
|
|
chr10_-_62373828
|
0.792
|
NM_014836
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chr14_+_101899104
|
0.786
|
|
TECPR2
|
tectonin beta-propeller repeat containing 2
|
|
chr3_-_187025438
|
0.785
|
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr19_-_18578467
|
0.784
|
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr10_+_128583984
|
0.782
|
NM_001380
|
DOCK1
|
dedicator of cytokinesis 1
|
|
chr12_-_123023052
|
0.779
|
NM_025140
|
CCDC92
|
coiled-coil domain containing 92
|
|
chr1_+_78217510
|
0.778
|
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4
|
|
chr20_+_11819364
|
0.776
|
NM_181443
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr15_+_61122139
|
0.775
|
|
TPM1
|
tropomyosin 1 (alpha)
|
|
chr2_-_16710551
|
0.773
|
NM_030797
|
FAM49A
|
family with sequence similarity 49, member A
|
|
chrX_-_17789276
|
0.769
|
NM_001172732
NM_001172739
NM_001172743
NM_021785
|
RAI2
|
retinoic acid induced 2
|
|
chr13_+_26029799
|
0.766
|
NM_006646
|
WASF3
|
WAS protein family, member 3
|
|
chr17_+_35853177
|
0.755
|
NM_001552
|
IGFBP4
|
insulin-like growth factor binding protein 4
|
|
chr5_-_126393967
|
0.753
|
|
MARCH3
|
membrane-associated ring finger (C3HC4) 3
|
|
chr3_-_48105637
|
0.751
|
NM_001134364
NM_002375
NM_030885
|
MAP4
|
microtubule-associated protein 4
|
|
chr12_-_45759691
|
0.751
|
NM_001143668
NM_181847
|
AMIGO2
|
adhesion molecule with Ig-like domain 2
|
|
chr2_-_73193419
|
0.751
|
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I)
|
|
chr17_-_60088637
|
0.750
|
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2
|
|
chr17_-_72218535
|
0.747
|
NM_001008528
NM_001008529
NM_198530
|
MXRA7
|
matrix-remodelling associated 7
|