|
chr10_+_53744043
|
5.377
|
NM_012242
|
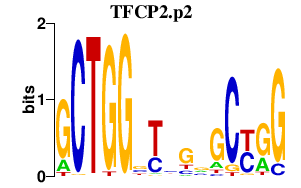
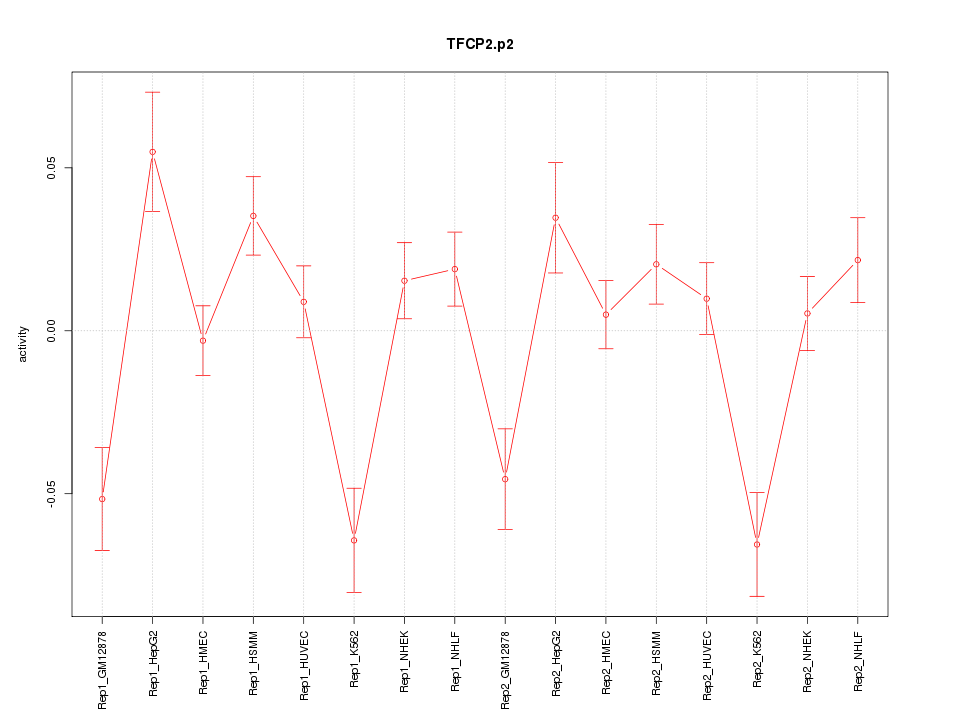
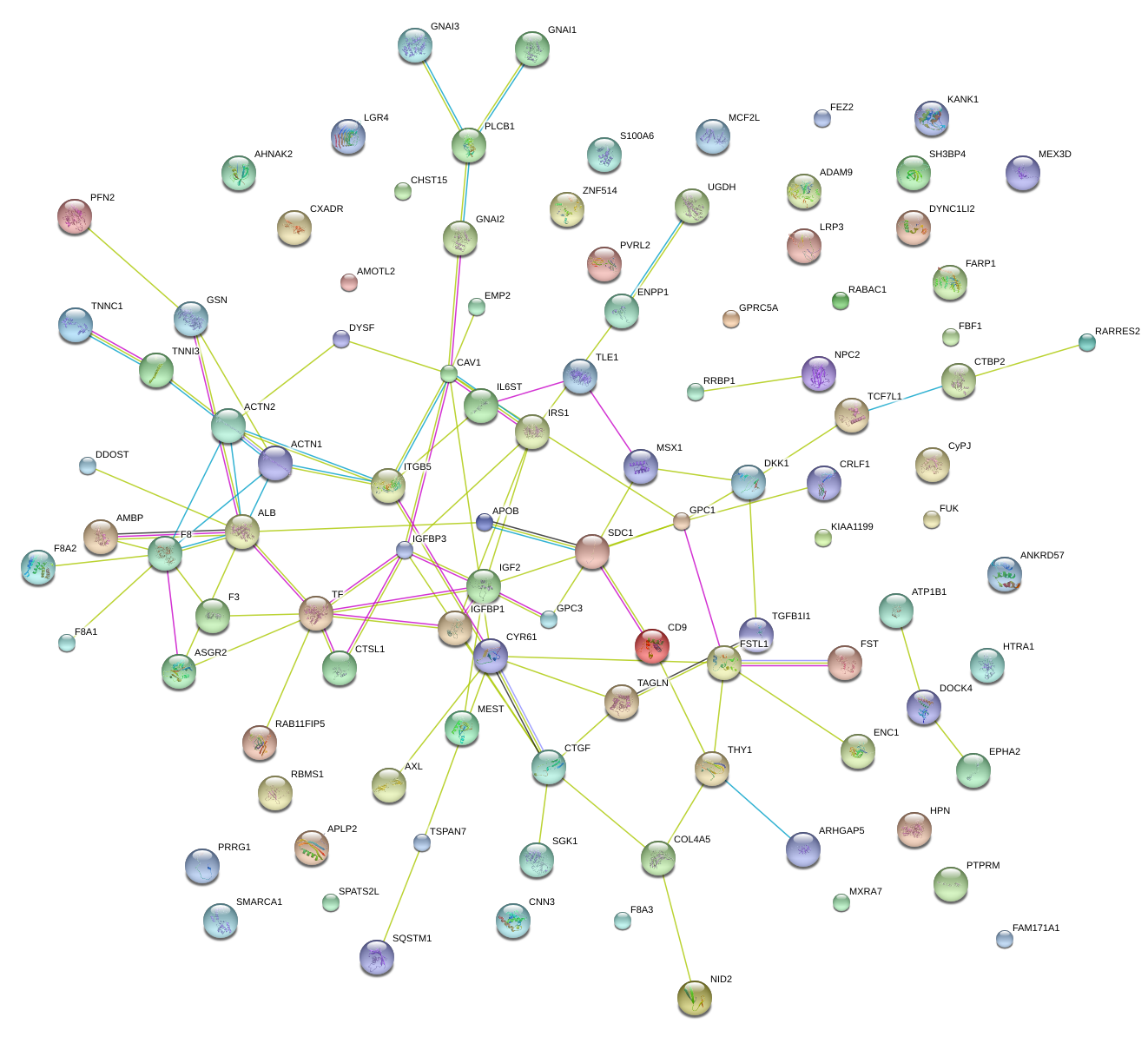
DKK1
|
dickkopf homolog 1 (Xenopus laevis)
|
|
chr2_-_161058120
|
5.292
|
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr10_+_124210980
|
4.563
|
NM_002775
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr2_-_161058550
|
4.467
|
NM_002897
NM_016836
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr11_-_2116501
|
4.345
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr11_-_2116048
|
3.730
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr10_+_124211353
|
3.497
|
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr11_-_2116360
|
3.023
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr7_+_129919157
|
2.919
|
NM_002402
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr11_+_129444877
|
2.850
|
NM_001142276
NM_001142277
NM_001142278
NM_001642
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr14_-_51605487
|
2.825
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr14_-_68515791
|
2.799
|
NM_001102
NM_001130004
NM_001130005
|
ACTN1
|
actinin, alpha 1
|
|
chr14_-_68515731
|
2.790
|
|
ACTN1
|
actinin, alpha 1
|
|
chr3_-_121652183
|
2.607
|
NM_007085
|
FSTL1
|
follistatin-like 1
|
|
chr15_+_78858763
|
2.589
|
NM_018689
|
KIAA1199
|
KIAA1199
|
|
chr20_-_17610828
|
2.578
|
NM_001042576
NM_004587
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr1_-_16355013
|
2.566
|
NM_004431
|
EPHA2
|
EPH receptor A2
|
|
chrX_-_132947290
|
2.565
|
NM_001164617
NM_001164618
NM_001164619
NM_004484
|
GPC3
|
glypican 3
|
|
chr14_-_51605515
|
2.478
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr19_+_38376980
|
2.460
|
|
LRP3
|
low density lipoprotein receptor-related protein 3
|
|
chr17_-_6958773
|
2.383
|
NM_001181
NM_080912
NM_080913
|
ASGR2
|
asialoglycoprotein receptor 2
|
|
chr1_-_20860524
|
2.381
|
|
DDOST
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase
|
|
chr14_-_51605461
|
2.346
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr14_-_68515761
|
2.343
|
|
ACTN1
|
actinin, alpha 1
|
|
chrX_-_132947141
|
2.319
|
|
GPC3
|
glypican 3
|
|
chr14_-_68515701
|
2.286
|
|
ACTN1
|
actinin, alpha 1
|
|
chr7_+_79602023
|
2.259
|
NM_002069
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1
|
|
chr11_-_2116775
|
2.250
|
NM_000612
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr19_+_46416938
|
2.220
|
NM_001699
NM_021913
|
AXL
|
AXL receptor tyrosine kinase
|
|
chr14_-_68515742
|
2.212
|
|
ACTN1
|
actinin, alpha 1
|
|
chr18_+_7557313
|
2.203
|
NM_001105244
NM_002845
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chrX_-_128485121
|
2.174
|
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chr3_+_134947573
|
2.142
|
NM_001063
|
TF
|
transferrin
|
|
chr2_+_200879656
|
2.133
|
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like
|
|
chr11_-_27450808
|
2.098
|
NM_018490
|
LGR4
|
leucine-rich repeat containing G protein-coupled receptor 4
|
|
chr1_-_95165089
|
2.097
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr10_-_126839546
|
2.069
|
NM_001083914
|
CTBP2
|
C-terminal binding protein 2
|
|
chr9_+_123070234
|
2.064
|
|
GSN
|
gelsolin
|
|
chrX_+_38305634
|
2.052
|
NM_004615
|
TSPAN7
|
tetraspanin 7
|
|
chr4_-_39205494
|
2.031
|
NM_001184700
NM_001184701
NM_003359
|
UGDH
|
UDP-glucose 6-dehydrogenase
|
|
chr11_+_129445002
|
2.029
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr17_-_6958695
|
2.013
|
|
ASGR2
|
asialoglycoprotein receptor 2
|
|
chr2_+_235525355
|
1.997
|
NM_014521
|
SH3BP4
|
SH3-domain binding protein 4
|
|
chr13_+_112681626
|
1.986
|
NM_024979
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr2_-_227371698
|
1.982
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr22_-_34754343
|
1.982
|
NM_001082578
NM_001082579
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr2_+_200879737
|
1.969
|
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like
|
|
chr20_+_8060762
|
1.941
|
NM_015192
NM_182734
|
PLCB1
|
phospholipase C, beta 1 (phosphoinositide-specific)
|
|
chr1_-_95165037
|
1.903
|
|
CNN3
|
calponin 3, acidic
|
|
chr3_+_134947940
|
1.868
|
|
TF
|
transferrin
|
|
chrX_+_153767803
|
1.833
|
NM_001007523
NM_001007524
NM_012151
|
F8A2
F8A3
F8A1
|
coagulation factor VIII-associated (intronic transcript) 2
coagulation factor VIII-associated (intronic transcript) 3
coagulation factor VIII-associated (intronic transcript) 1
|
|
chr9_+_494661
|
1.822
|
NM_015158
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr3_-_151171190
|
1.802
|
NM_002628
NM_053024
|
PFN2
|
profilin 2
|
|
chr5_+_52812165
|
1.777
|
|
FST
|
follistatin
|
|
chrX_-_154341451
|
1.750
|
NM_001007523
NM_001007524
NM_012151
|
F8A2
F8A3
F8A1
|
coagulation factor VIII-associated (intronic transcript) 2
coagulation factor VIII-associated (intronic transcript) 3
coagulation factor VIII-associated (intronic transcript) 1
|
|
chr2_-_95189231
|
1.718
|
|
ZNF514
|
zinc finger protein 514
|
|
chr4_-_39205316
|
1.717
|
|
UGDH
|
UDP-glucose 6-dehydrogenase
|
|
chr14_-_68515525
|
1.714
|
|
ACTN1
|
actinin, alpha 1
|
|
chr5_-_73972055
|
1.707
|
|
ENC1
|
ectodermal-neural cortex 1 (with BTB-like domain)
|
|
chr19_-_47155277
|
1.698
|
|
RABAC1
|
Rab acceptor 1 (prenylated)
|
|
chr19_+_38377329
|
1.670
|
NM_002333
|
LRP3
|
low density lipoprotein receptor-related protein 3
|
|
chr14_-_51605695
|
1.665
|
NM_007361
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr11_+_129445057
|
1.660
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr7_-_149669622
|
1.644
|
NM_002889
|
RARRES2
|
retinoic acid receptor responder (tazarotene induced) 2
|
|
chr11_+_129444997
|
1.640
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr9_+_123070191
|
1.636
|
NM_001127662
NM_001127663
NM_001127664
NM_001127665
NM_001127666
NM_001127667
NM_198252
|
GSN
|
gelsolin
|
|
chr6_-_134538526
|
1.635
|
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr5_+_52812236
|
1.630
|
NM_006350
NM_013409
|
FST
|
follistatin
|
|
chr2_-_20288108
|
1.612
|
NM_002997
|
SDC1
|
syndecan 1
|
|
chr7_+_115952325
|
1.612
|
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr7_+_45894480
|
1.603
|
NM_000596
|
IGFBP1
|
insulin-like growth factor binding protein 1
|
|
chr2_-_73193419
|
1.585
|
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I)
|
|
chr3_-_135575925
|
1.557
|
NM_016201
|
AMOTL2
|
angiomotin like 2
|
|
chr19_-_18578577
|
1.548
|
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr7_+_115952279
|
1.547
|
NM_001172895
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr17_-_72218661
|
1.538
|
|
MXRA7
|
matrix-remodelling associated 7
|
|
chr19_-_47155340
|
1.538
|
NM_006423
|
RABAC1
|
Rab acceptor 1 (prenylated)
|
|
chr12_+_6179736
|
1.533
|
NM_001769
|
CD9
|
CD9 molecule
|
|
chr3_-_52463070
|
1.507
|
NM_003280
|
TNNC1
|
troponin C type 1 (slow)
|
|
chrX_-_128485062
|
1.486
|
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chr16_+_69060325
|
1.480
|
|
FUK
|
fucokinase
|
|
chr20_-_17610704
|
1.471
|
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr16_-_10581944
|
1.459
|
NM_001424
|
EMP2
|
epithelial membrane protein 2
|
|
chr9_+_123003549
|
1.446
|
|
GSN
|
gelsolin
|
|
chrX_-_128485134
|
1.423
|
NM_003069
NM_139035
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chr9_-_115877104
|
1.410
|
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr14_-_74029788
|
1.406
|
NM_006432
|
NPC2
|
Niemann-Pick disease, type C2
|
|
chr14_-_104515738
|
1.401
|
NM_138420
|
AHNAK2
|
AHNAK nucleoprotein 2
|
|
chr10_-_125841840
|
1.399
|
NM_014863
NM_015892
|
CHST15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15
|
|
chr16_+_31390567
|
1.387
|
NM_001042454
NM_001164719
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr6_+_132170888
|
1.380
|
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chr19_+_50041232
|
1.371
|
NM_001042724
NM_002856
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr19_+_50041400
|
1.365
|
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr9_+_89530943
|
1.357
|
|
CTSL1
|
cathepsin L1
|
|
chrX_+_107569671
|
1.356
|
NM_000495
NM_033380
|
COL4A5
|
collagen, type IV, alpha 5
|
|
chr12_+_12935619
|
1.355
|
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr14_-_74029756
|
1.354
|
|
NPC2
|
Niemann-Pick disease, type C2
|
|
chr19_+_50041387
|
1.353
|
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr17_-_6958282
|
1.352
|
NM_080914
|
ASGR2
|
asialoglycoprotein receptor 2
|
|
chr4_+_4912272
|
1.350
|
NM_002448
|
MSX1
|
msh homeobox 1
|
|
chr4_+_74496660
|
1.348
|
|
ALB
|
albumin
|
|
chr12_+_12935222
|
1.347
|
NM_003979
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr3_-_126088819
|
1.346
|
NM_002213
|
ITGB5
|
integrin, beta 5
|
|
chr4_+_74496575
|
1.343
|
|
ALB
|
albumin
|
|
chr2_+_71534260
|
1.340
|
NM_001130976
NM_001130977
NM_001130978
NM_001130979
NM_001130980
NM_001130981
NM_003494
|
DYSF
|
dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive)
|
|
chr7_-_111633620
|
1.333
|
NM_014705
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr3_-_126088530
|
1.314
|
|
ITGB5
|
integrin, beta 5
|
|
chr14_-_74029720
|
1.308
|
|
NPC2
|
Niemann-Pick disease, type C2
|
|
chr11_+_116575249
|
1.308
|
NM_001001522
NM_003186
|
TAGLN
|
transgelin
|
|
chr1_+_85819004
|
1.307
|
NM_001554
|
CYR61
|
cysteine-rich, angiogenic inducer, 61
|
|
chr1_-_151775087
|
1.306
|
|
S100A6
|
S100 calcium binding protein A6
|
|
chr2_+_109729176
|
1.303
|
NM_023016
|
ANKRD57
|
ankyrin repeat domain 57
|
|
chr2_-_95188989
|
1.301
|
NM_032788
|
ZNF514
|
zinc finger protein 514
|
|
chr8_+_38973681
|
1.300
|
|
ADAM9
|
ADAM metallopeptidase domain 9
|
|
chr7_+_129918408
|
1.287
|
NM_177525
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr10_-_15453060
|
1.283
|
NM_001010924
|
FAM171A1
|
family with sequence similarity 171, member A1
|
|
chr1_+_167342493
|
1.280
|
NM_001677
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr9_-_83494198
|
1.274
|
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chrX_+_37093448
|
1.273
|
NM_000950
NM_001142395
NM_001173486
NM_001173489
NM_001173490
|
PRRG1
|
proline rich Gla (G-carboxyglutamic acid) 1
|
|
chr14_-_68515727
|
1.270
|
|
ACTN1
|
actinin, alpha 1
|
|
chr11_-_118799452
|
1.268
|
NM_006288
|
THY1
|
Thy-1 cell surface antigen
|
|
chr11_+_129445017
|
1.268
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr19_+_40223249
|
1.261
|
NM_002151
NM_182983
|
HPN
|
hepsin
|
|
chr14_+_31616196
|
1.252
|
NM_001030055
NM_001173
|
ARHGAP5
|
Rho GTPase activating protein 5
|
|
chr13_+_97593711
|
1.248
|
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr16_-_65343012
|
1.248
|
NM_006141
|
DYNC1LI2
|
dynein, cytoplasmic 1, light intermediate chain 2
|
|
chr5_-_55326358
|
1.247
|
NM_001190981
NM_002184
NM_175767
|
IL6ST
|
interleukin 6 signal transducer (gp130, oncostatin M receptor)
|
|
chr2_-_36678657
|
1.245
|
NM_001042548
NM_005102
|
FEZ2
|
fasciculation and elongation protein zeta 2 (zygin II)
|
|
chr2_-_20288647
|
1.218
|
NM_001006946
|
SDC1
|
syndecan 1
|
|
chr6_-_132314004
|
1.213
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr21_+_17807167
|
1.200
|
NM_001338
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr2_+_85213884
|
1.200
|
NM_031283
|
TCF7L1
|
transcription factor 7-like 1 (T-cell specific, HMG-box)
|
|
chr19_-_1518875
|
1.199
|
NM_001174118
NM_203304
|
MEX3D
|
mex-3 homolog D (C. elegans)
|
|
chr5_+_179166608
|
1.193
|
NM_001142299
|
SQSTM1
|
sequestosome 1
|
|
chr2_+_241023703
|
1.191
|
NM_002081
|
GPC1
|
glypican 1
|
|
chr2_-_21120398
|
1.189
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr7_-_45927263
|
1.187
|
NM_000598
NM_001013398
|
IGFBP3
|
insulin-like growth factor binding protein 3
|
|
chr16_+_55199978
|
1.184
|
NM_005953
|
MT2A
|
metallothionein 2A
|
|
chr12_+_6923441
|
1.182
|
NM_138425
|
C12orf57
|
chromosome 12 open reading frame 57
|
|
chr14_+_104402626
|
1.182
|
NM_001112726
NM_015005
|
KIAA0284
|
KIAA0284
|
|
chr2_-_21120314
|
1.170
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr1_-_9806611
|
1.165
|
|
CLSTN1
|
calsyntenin 1
|
|
chr17_+_21128625
|
1.159
|
|
MAP2K3
|
mitogen-activated protein kinase kinase 3
|
|
chr13_+_97593469
|
1.158
|
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr12_+_12935759
|
1.157
|
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr10_-_91001566
|
1.153
|
NM_000235
NM_001127605
|
LIPA
|
lipase A, lysosomal acid, cholesterol esterase
|
|
chr1_-_151784889
|
1.152
|
NM_002961
NM_019554
|
S100A4
|
S100 calcium binding protein A4
|
|
chr13_+_97593411
|
1.151
|
NM_001001715
NM_005766
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr2_-_21120420
|
1.148
|
NM_000384
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr10_-_91001496
|
1.148
|
|
LIPA
|
lipase A, lysosomal acid, cholesterol esterase
|
|
chr16_+_56211150
|
1.143
|
|
GPR56
|
G protein-coupled receptor 56
|
|
chr10_-_91001531
|
1.141
|
|
LIPA
|
lipase A, lysosomal acid, cholesterol esterase
|
|
chr22_+_36531246
|
1.141
|
|
H1F0
|
H1 histone family, member 0
|
|
chr3_+_180853473
|
1.132
|
NM_003940
|
USP13
|
ubiquitin specific peptidase 13 (isopeptidase T-3)
|
|
chr16_+_8799170
|
1.127
|
NM_000303
|
PMM2
|
phosphomannomutase 2
|
|
chr10_-_91001542
|
1.116
|
|
LIPA
|
lipase A, lysosomal acid, cholesterol esterase
|
|
chr14_+_61231795
|
1.112
|
NM_001530
NM_181054
|
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor)
|
|
chr16_+_31391998
|
1.107
|
NM_015927
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr1_-_151775119
|
1.106
|
|
S100A6
|
S100 calcium binding protein A6
|
|
chr17_+_21128560
|
1.106
|
NM_145109
|
MAP2K3
|
mitogen-activated protein kinase kinase 3
|
|
chr1_-_150232927
|
1.104
|
|
S100A10
|
S100 calcium binding protein A10
|
|
chr7_+_65307816
|
1.103
|
|
TPST1
|
tyrosylprotein sulfotransferase 1
|
|
chr6_+_146906520
|
1.099
|
NM_006834
|
RAB32
|
RAB32, member RAS oncogene family
|
|
chr19_+_41298177
|
1.097
|
|
TBCB
|
tubulin folding cofactor B
|
|
chr1_-_151775221
|
1.096
|
NM_014624
|
S100A6
|
S100 calcium binding protein A6
|
|
chr10_+_123913094
|
1.081
|
NM_006997
NM_206860
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr7_+_17304707
|
1.080
|
NM_001621
|
AHR
|
aryl hydrocarbon receptor
|
|
chr7_+_65307691
|
1.077
|
NM_003596
|
TPST1
|
tyrosylprotein sulfotransferase 1
|
|
chr10_-_93382837
|
1.077
|
NM_005398
|
PPP1R3C
|
protein phosphatase 1, regulatory (inhibitor) subunit 3C
|
|
chr9_-_109291866
|
1.072
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr4_-_1156533
|
1.067
|
NM_012445
|
SPON2
|
spondin 2, extracellular matrix protein
|
|
chr6_-_134538762
|
1.066
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr2_-_73193581
|
1.064
|
NM_015470
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I)
|
|
chr10_+_123912930
|
1.061
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr12_-_123023312
|
1.037
|
|
CCDC92
|
coiled-coil domain containing 92
|
|
chr4_-_1156458
|
1.037
|
|
SPON2
|
spondin 2, extracellular matrix protein
|
|
chr2_-_24160683
|
1.036
|
|
TP53I3
|
tumor protein p53 inducible protein 3
|
|
chr8_-_48813256
|
1.031
|
NM_005195
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta
|
|
chrX_+_43399101
|
1.030
|
|
MAOA
|
monoamine oxidase A
|
|
chr16_-_65342979
|
1.018
|
|
DYNC1LI2
|
dynein, cytoplasmic 1, light intermediate chain 2
|
|
chr17_+_45858517
|
1.008
|
NM_025149
|
ACSF2
|
acyl-CoA synthetase family member 2
|
|
chr2_+_9264327
|
1.004
|
NM_001135191
NM_003887
|
ASAP2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2
|
|
chr10_+_102780974
|
1.002
|
NM_030971
|
SFXN3
|
sideroflexin 3
|
|
chrX_-_106130098
|
0.999
|
NM_001085354
NM_024657
|
MORC4
|
MORC family CW-type zinc finger 4
|
|
chr20_+_44771532
|
0.997
|
NM_030777
|
SLC2A10
|
solute carrier family 2 (facilitated glucose transporter), member 10
|
|
chr6_+_146906658
|
0.995
|
|
RAB32
|
RAB32, member RAS oncogene family
|
|
chr5_-_179166378
|
0.993
|
NM_014275
|
MGAT4B
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B
|
|
chr2_+_9264360
|
0.992
|
|
ASAP2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2
|
|
chr17_+_1611983
|
0.990
|
NM_002615
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1
|
|
chr22_-_49063088
|
0.986
|
|
PLXNB2
|
plexin B2
|
|
chr4_+_6627816
|
0.981
|
|
MAN2B2
|
mannosidase, alpha, class 2B, member 2
|
|
chr6_-_74420410
|
0.981
|
NM_012434
|
SLC17A5
|
solute carrier family 17 (anion/sugar transporter), member 5
|
|
chr6_-_74420380
|
0.981
|
|
SLC17A5
|
solute carrier family 17 (anion/sugar transporter), member 5
|
|
chr7_+_93861808
|
0.963
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr9_-_73573121
|
0.960
|
NM_001135820
NM_013390
|
TMEM2
|
transmembrane protein 2
|
|
chr17_+_8279860
|
0.954
|
NM_001025579
NM_030808
|
NDEL1
|
nudE nuclear distribution gene E homolog (A. nidulans)-like 1
|
|
chr6_-_134540666
|
0.954
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr1_+_17779528
|
0.953
|
NM_001011722
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like
|
|
chr17_+_1612056
|
0.953
|
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1
|
|
chr7_-_29995809
|
0.949
|
NM_001145515
NM_014766
NM_001145513
|
SCRN1
|
secernin 1
|