|
chr13_+_112999538
|
15.229
|
|
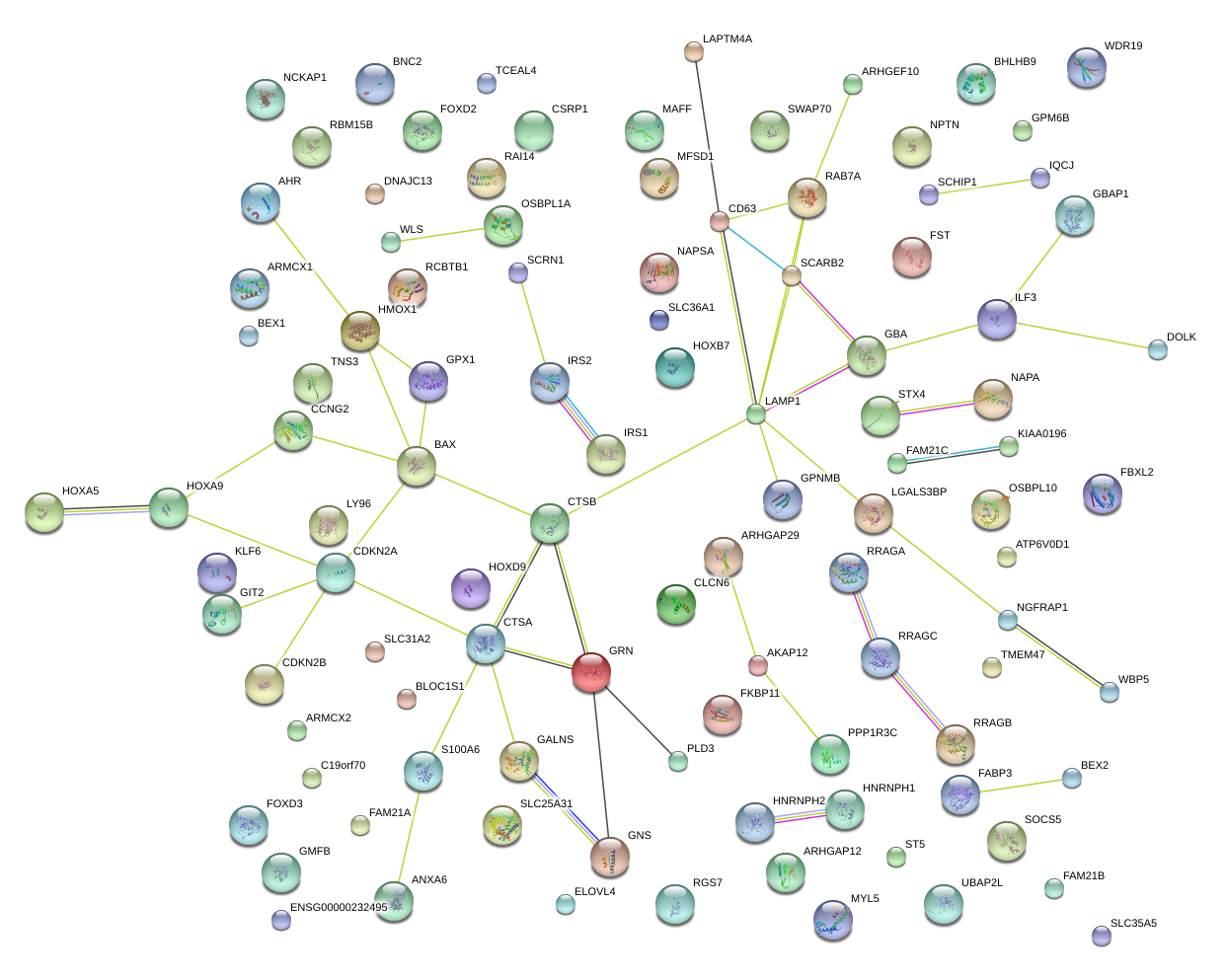
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr13_+_112999462
|
13.625
|
NM_005561
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr13_+_112999590
|
11.682
|
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chrX_+_102518064
|
10.940
|
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr19_+_54149843
|
9.599
|
NM_004324
NM_138761
NM_138763
NM_138764
|
BAX
|
BCL2-associated X protein
|
|
chr19_+_54149941
|
9.590
|
|
BAX
|
BCL2-associated X protein
|
|
chr19_+_54149963
|
8.814
|
|
BAX
|
BCL2-associated X protein
|
|
chr1_-_151775221
|
8.473
|
NM_014624
|
S100A6
|
S100 calcium binding protein A6
|
|
chr1_-_68470585
|
7.697
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chr3_-_49370755
|
7.669
|
NM_000581
NM_201397
|
GPX1
|
glutathione peroxidase 1
|
|
chr20_+_43953428
|
7.590
|
|
CTSA
|
cathepsin A
|
|
chr20_+_43953372
|
7.451
|
|
CTSA
|
cathepsin A
|
|
chrX_-_100801472
|
7.442
|
NM_014782
NM_177949
|
ARMCX2
|
armadillo repeat containing, X-linked 2
|
|
chr3_-_49370682
|
7.437
|
|
GPX1
|
glutathione peroxidase 1
|
|
chr5_+_34692124
|
7.389
|
NM_001145522
NM_015577
|
RAI14
|
retinoic acid induced 14
|
|
chr22_+_34107087
|
7.370
|
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr7_-_47588652
|
7.189
|
|
TNS3
|
tensin 3
|
|
chr15_-_71712711
|
7.125
|
NM_001161363
NM_001161364
NM_012428
NM_017455
|
NPTN
|
neuroplastin
|
|
chr22_+_34107051
|
7.054
|
NM_002133
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr12_-_47604930
|
6.889
|
NM_001143781
|
FKBP11
|
FK506 binding protein 11, 19 kDa
|
|
chrX_-_34585287
|
6.831
|
NM_031442
|
TMEM47
|
transmembrane protein 47
|
|
chr19_-_10625449
|
6.785
|
|
LOC147727
|
hypothetical LOC147727
|
|
chr1_-_151775087
|
6.717
|
|
S100A6
|
S100 calcium binding protein A6
|
|
chr6_+_151603201
|
6.563
|
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr5_+_52812165
|
6.495
|
|
FST
|
follistatin
|
|
chr5_+_34692239
|
6.350
|
|
RAI14
|
retinoic acid induced 14
|
|
chr6_+_151602787
|
6.293
|
NM_005100
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr7_-_47588160
|
6.222
|
|
TNS3
|
tensin 3
|
|
chr15_-_71712605
|
6.068
|
|
NPTN
|
neuroplastin
|
|
chr1_-_153477511
|
6.068
|
|
GBA
|
glucosidase, beta, acid
|
|
chr1_-_151775119
|
6.020
|
|
S100A6
|
S100 calcium binding protein A6
|
|
chr20_+_43953606
|
6.017
|
|
CTSA
|
cathepsin A
|
|
chr1_-_153477673
|
5.848
|
NM_001171812
NM_000157
|
GBA
|
glucosidase, beta, acid
|
|
chr20_+_43953331
|
5.776
|
|
CTSA
|
cathepsin A
|
|
chr1_-_94475840
|
5.746
|
NM_004815
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr7_-_47588266
|
5.702
|
|
TNS3
|
tensin 3
|
|
chr5_+_34692352
|
5.638
|
NM_001145520
|
RAI14
|
retinoic acid induced 14
|
|
chr12_-_63439456
|
5.357
|
NM_002076
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr2_+_46779553
|
5.303
|
NM_014011
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr1_+_11788827
|
5.241
|
|
CLCN6
|
chloride channel 6
|
|
chrX_+_100692071
|
5.235
|
NM_016608
|
ARMCX1
|
armadillo repeat containing, X-linked 1
|
|
chr3_+_113763561
|
5.151
|
NM_017945
|
SLC35A5
|
solute carrier family 35, member A5
|
|
chr9_+_114953048
|
4.991
|
NM_001860
|
SLC31A2
|
solute carrier family 31 (copper transporters), member 2
|
|
chr17_+_39778016
|
4.982
|
NM_002087
|
GRN
|
granulin
|
|
chr10_-_32257768
|
4.953
|
NM_018287
|
ARHGAP12
|
Rho GTPase activating protein 12
|
|
chr5_+_52812236
|
4.882
|
NM_006350
NM_013409
|
FST
|
follistatin
|
|
chr2_-_227372574
|
4.799
|
|
IRS1
|
insulin receptor substrate 1
|
|
chr3_+_160002605
|
4.722
|
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chr10_-_32257693
|
4.681
|
|
ARHGAP12
|
Rho GTPase activating protein 12
|
|
chr1_+_152459918
|
4.585
|
NM_014847
|
UBAP2L
|
ubiquitin associated protein 2-like
|
|
chr3_+_33293915
|
4.565
|
NM_012157
NM_001171713
|
FBXL2
|
F-box and leucine-rich repeat protein 2
|
|
chr17_+_39778153
|
4.412
|
|
GRN
|
granulin
|
|
chr9_+_114953087
|
4.283
|
|
SLC31A2
|
solute carrier family 31 (copper transporters), member 2
|
|
chr16_-_66072477
|
4.276
|
NM_004691
|
ATP6V0D1
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1
|
|
chr17_+_39778191
|
4.231
|
|
GRN
|
granulin
|
|
chr7_-_27149750
|
4.228
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr12_-_63439449
|
4.228
|
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr3_+_160002571
|
4.154
|
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chr7_+_23252909
|
4.129
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr17_+_39778180
|
4.063
|
|
GRN
|
granulin
|
|
chr3_+_51403760
|
4.032
|
NM_013286
|
RBM15B
|
RNA binding motif protein 15B
|
|
chr3_+_113763601
|
4.030
|
|
SLC35A5
|
solute carrier family 35, member A5
|
|
chr3_+_160473729
|
3.986
|
NM_001197107
NM_001197108
NM_014575
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr16_-_87450849
|
3.982
|
NM_000512
|
GALNS
|
galactosamine (N-acetyl)-6-sulfate sulfatase
|
|
chr12_-_54409107
|
3.976
|
NM_001040034
NM_001780
|
CD63
|
CD63 molecule
|
|
chr7_-_29995809
|
3.929
|
NM_001145515
NM_014766
NM_001145513
|
SCRN1
|
secernin 1
|
|
chr4_+_78297356
|
3.917
|
NM_004354
|
CCNG2
|
cyclin G2
|
|
chr6_+_151603346
|
3.909
|
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr19_+_45546369
|
3.881
|
|
PLD3
|
phospholipase D family, member 3
|
|
chr3_+_129927716
|
3.874
|
|
RAB7A
|
RAB7A, member RAS oncogene family
|
|
chr6_+_151688327
|
3.842
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chrX_+_102727068
|
3.832
|
NM_001006935
NM_001006937
NM_024863
|
TCEAL4
|
transcription elongation factor A (SII)-like 4
|
|
chr3_+_160002619
|
3.824
|
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chrX_+_55760774
|
3.785
|
NM_006064
NM_016656
|
RRAGB
|
Ras-related GTP binding B
|
|
chr1_-_94475504
|
3.762
|
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr3_+_133619193
|
3.749
|
NM_015268
|
DNAJC13
|
DnaJ (Hsp40) homolog, subfamily C, member 13
|
|
chr6_-_80713589
|
3.738
|
NM_022726
|
ELOVL4
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4
|
|
chr17_-_44043361
|
3.725
|
NM_004502
|
HOXB7
|
homeobox B7
|
|
chr7_+_23252915
|
3.693
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr8_-_126173189
|
3.679
|
|
KIAA0196
|
KIAA0196
|
|
chr1_-_39097904
|
3.678
|
NM_022157
|
RRAGC
|
Ras-related GTP binding C
|
|
chr9_-_16860719
|
3.640
|
NM_017637
|
BNC2
|
basonuclin 2
|
|
chr16_-_87450734
|
3.618
|
|
GALNS
|
galactosamine (N-acetyl)-6-sulfate sulfatase
|
|
chr8_+_1759502
|
3.594
|
NM_014629
|
ARHGEF10
|
Rho guanine nucleotide exchange factor (GEF) 10
|
|
chr3_-_31998241
|
3.579
|
NM_001174060
NM_017784
|
OSBPL10
|
oxysterol binding protein-like 10
|
|
chr8_-_11763037
|
3.542
|
NM_001908
NM_147780
NM_147781
NM_147782
NM_147783
|
CTSB
|
cathepsin B
|
|
chr3_+_129927723
|
3.541
|
|
RAB7A
|
RAB7A, member RAS oncogene family
|
|
chr7_+_23252911
|
3.526
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr12_-_54409059
|
3.520
|
|
CD63
|
CD63 molecule
|
|
chr5_+_150807331
|
3.519
|
NM_078483
|
SLC36A1
|
solute carrier family 36 (proton/amino acid symporter), member 1
|
|
chr12_-_54409015
|
3.516
|
|
CD63
|
CD63 molecule
|
|
chr7_+_17304707
|
3.480
|
NM_001621
|
AHR
|
aryl hydrocarbon receptor
|
|
chr8_-_126173055
|
3.475
|
|
KIAA0196
|
KIAA0196
|
|
chr3_+_129927668
|
3.471
|
NM_004637
|
RAB7A
|
RAB7A, member RAS oncogene family
|
|
chr22_+_36928972
|
3.466
|
NM_001161573
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr8_-_126173186
|
3.451
|
|
KIAA0196
|
KIAA0196
|
|
chr16_+_30952382
|
3.416
|
NM_004604
|
STX4
|
syntaxin 4
|
|
chr13_-_49057629
|
3.412
|
|
RCBTB1
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1
|
|
chr10_-_93382837
|
3.412
|
NM_005398
|
PPP1R3C
|
protein phosphatase 1, regulatory (inhibitor) subunit 3C
|
|
chr10_-_3817418
|
3.405
|
NM_001160124
NM_001160125
NM_001300
|
KLF6
|
Kruppel-like factor 6
|
|
chr11_+_9642226
|
3.404
|
|
SWAP70
|
SWAP switching B-cell complex 70kDa subunit
|
|
chr4_-_77354011
|
3.385
|
NM_005506
|
SCARB2
|
scavenger receptor class B, member 2
|
|
chr19_-_52710129
|
3.381
|
NM_003827
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chr7_+_23252945
|
3.372
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr1_+_11788793
|
3.366
|
NM_001286
NM_021735
NM_021736
NM_021737
|
CLCN6
|
chloride channel 6
|
|
chr13_-_109236897
|
3.363
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr1_-_31618405
|
3.327
|
NM_004102
|
FABP3
|
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor)
|
|
chr17_-_74487527
|
3.318
|
NM_005567
|
LGALS3BP
|
lectin, galactoside-binding, soluble, 3 binding protein
|
|
chr5_-_72780107
|
3.297
|
NM_004472
|
FOXD1
|
forkhead box D1
|
|
chr3_+_129927714
|
3.285
|
|
RAB7A
|
RAB7A, member RAS oncogene family
|
|
chr19_+_10625896
|
3.269
|
NM_001137673
NM_004516
NM_012218
NM_017620
NM_153464
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr7_-_27171673
|
3.240
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chrX_+_101862297
|
3.231
|
NM_001142524
NM_001142525
NM_001142526
NM_001142527
NM_001142528
NM_030639
|
BHLHB9
|
basic helix-loop-helix domain containing, class B, 9
|
|
chr8_-_126173108
|
3.231
|
|
KIAA0196
|
KIAA0196
|
|
chr11_-_8849485
|
3.204
|
|
ST5
|
suppression of tumorigenicity 5
|
|
chr7_-_27171648
|
3.160
|
|
HOXA9
|
homeobox A9
|
|
chr16_+_30952350
|
3.157
|
|
STX4
|
syntaxin 4
|
|
chrX_+_100549861
|
3.130
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr12_-_54409067
|
3.113
|
|
CD63
|
CD63 molecule
|
|
chr8_-_126173203
|
3.100
|
|
KIAA0196
|
KIAA0196
|
|
chr11_+_9642200
|
3.061
|
NM_015055
|
SWAP70
|
SWAP switching B-cell complex 70kDa subunit
|
|
chr4_+_128870968
|
3.056
|
NM_031291
|
SLC25A31
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 31
|
|
chr9_-_21984414
|
3.036
|
NM_058195
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chr1_-_199742589
|
3.015
|
NM_001193571
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr12_-_108918411
|
2.984
|
|
GIT2
|
G protein-coupled receptor kinase interacting ArfGAP 2
|
|
chr14_-_54025414
|
2.982
|
|
GMFB
|
glia maturation factor, beta
|
|
chr19_-_5631910
|
2.977
|
NM_205767
|
C19orf70
|
chromosome 19 open reading frame 70
|
|
chr14_-_54025464
|
2.969
|
NM_004124
|
GMFB
|
glia maturation factor, beta
|
|
chr9_-_16860664
|
2.916
|
|
BNC2
|
basonuclin 2
|
|
chrX_+_102498035
|
2.905
|
NM_001006612
NM_001006613
NM_001006614
NM_016303
|
WBP5
|
WW domain binding protein 5
|
|
chrX_-_13866451
|
2.901
|
|
GPM6B
|
glycoprotein M6B
|
|
chrX_+_100549839
|
2.883
|
NM_001032393
NM_019597
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chrX_+_100549869
|
2.880
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr9_-_130749604
|
2.874
|
NM_014908
|
DOLK
|
dolichol kinase
|
|
chrX_-_102205736
|
2.868
|
|
BEX1
|
brain expressed, X-linked 1
|
|
chr4_+_38860418
|
2.855
|
NM_025132
|
WDR19
|
WD repeat domain 19
|
|
chrX_+_100549885
|
2.852
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr18_-_20231709
|
2.851
|
|
OSBPL1A
|
oxysterol binding protein-like 1A
|
|
chr8_+_75066117
|
2.836
|
NM_001195797
NM_015364
|
LY96
|
lymphocyte antigen 96
|
|
chr1_-_239587090
|
2.827
|
NM_002924
|
RGS7
|
regulator of G-protein signaling 7
|
|
chr2_-_183611470
|
2.786
|
NM_013436
NM_205842
|
NCKAP1
|
NCK-associated protein 1
|
|
chr12_+_54396099
|
2.783
|
|
BLOC1S1-RDH5
BLOC1S1
|
BLOC1S1-RDH5 read-through transcript
biogenesis of lysosomal organelles complex-1, subunit 1
|
|
chr4_+_658250
|
2.769
|
|
MYL5
|
myosin, light chain 5, regulatory
|
|
chr5_-_150517499
|
2.752
|
|
ANXA6
|
annexin A6
|
|
chr4_+_78297418
|
2.749
|
|
CCNG2
|
cyclin G2
|
|
chr2_-_20115269
|
2.745
|
NM_014713
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha
|
|
chr10_+_45542653
|
2.727
|
NM_001169106
NM_001169107
NM_015262
|
FAM21C
FAM21A
|
family with sequence similarity 21, member C
family with sequence similarity 21, member A
|
|
chr12_-_63439415
|
2.697
|
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr3_+_160002582
|
2.682
|
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chr17_+_18159294
|
2.656
|
NM_144775
|
SMCR8
|
Smith-Magenis syndrome chromosome region, candidate 8
|
|
chr1_-_239587007
|
2.644
|
|
RGS7
|
regulator of G-protein signaling 7
|
|
chr13_-_49057707
|
2.641
|
NM_018191
|
RCBTB1
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1
|
|
chr17_-_7078446
|
2.639
|
NM_004422
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr10_+_51497716
|
2.620
|
|
FAM21A
|
family with sequence similarity 21, member A
|
|
chr5_-_172687798
|
2.618
|
|
STC2
|
stanniocalcin 2
|
|
chr14_-_91642703
|
2.603
|
NM_001127696
NM_001127697
NM_001164774
NM_001164776
NM_001164777
NM_001164778
NM_001164779
NM_001164780
NM_001164781
NM_001164782
NM_004993
NM_030660
|
ATXN3
|
ataxin 3
|
|
chr10_-_3817205
|
2.585
|
|
KLF6
|
Kruppel-like factor 6
|
|
chr8_-_126173227
|
2.574
|
NM_014846
|
KIAA0196
|
KIAA0196
|
|
chr12_+_6703686
|
2.558
|
NM_001164093
|
COPS7A
|
COP9 constitutive photomorphogenic homolog subunit 7A (Arabidopsis)
|
|
chrX_+_102770547
|
2.537
|
NM_001006639
NM_004780
|
TCEAL1
|
transcription elongation factor A (SII)-like 1
|
|
chr4_+_658312
|
2.500
|
|
MYL5
|
myosin, light chain 5, regulatory
|
|
chr20_+_56318245
|
2.499
|
|
RAB22A
|
RAB22A, member RAS oncogene family
|
|
chr8_-_95343696
|
2.495
|
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr11_+_101485946
|
2.490
|
NM_001130145
NM_001195044
NM_006106
|
YAP1
|
Yes-associated protein 1
|
|
chrX_+_100765006
|
2.480
|
NM_177948
|
ARMCX3
|
armadillo repeat containing, X-linked 3
|
|
chr5_-_172130766
|
2.474
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chrX_-_106846924
|
2.454
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr14_-_68515791
|
2.422
|
NM_001102
NM_001130004
NM_001130005
|
ACTN1
|
actinin, alpha 1
|
|
chr11_-_27679617
|
2.418
|
NM_170733
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_1741680
|
2.417
|
|
CTSD
|
cathepsin D
|
|
chr3_+_160002408
|
2.351
|
NM_001167903
NM_022736
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chr2_+_238060602
|
2.349
|
NM_001042467
NM_024101
|
MLPH
|
melanophilin
|
|
chr6_-_134538762
|
2.344
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr14_-_67353046
|
2.340
|
NM_015346
|
ZFYVE26
|
zinc finger, FYVE domain containing 26
|
|
chr8_-_95343716
|
2.327
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr2_+_223625105
|
2.323
|
NM_080671
|
KCNE4
|
potassium voltage-gated channel, Isk-related family, member 4
|
|
chr5_+_149320480
|
2.322
|
NM_000112
|
SLC26A2
|
solute carrier family 26 (sulfate transporter), member 2
|
|
chr5_+_172343415
|
2.321
|
|
ATP6V0E1
|
ATPase, H+ transporting, lysosomal 9kDa, V0 subunit e1
|
|
chr8_+_26296439
|
2.318
|
NM_004331
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like
|
|
chr13_-_95094920
|
2.316
|
NM_014934
NM_198968
|
DZIP1
|
DAZ interacting protein 1
|
|
chr9_-_21984379
|
2.314
|
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chr20_+_56318155
|
2.309
|
NM_020673
|
RAB22A
|
RAB22A, member RAS oncogene family
|
|
chrX_-_102205752
|
2.253
|
NM_018476
|
BEX1
|
brain expressed, X-linked 1
|
|
chr11_-_1741663
|
2.238
|
|
CTSD
|
cathepsin D
|
|
chrX_+_100764757
|
2.233
|
NM_016607
NM_177947
|
ARMCX3
|
armadillo repeat containing, X-linked 3
|
|
chr12_+_64504408
|
2.228
|
NM_003483
NM_003484
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr10_+_51497660
|
2.227
|
NM_001005751
|
FAM21C
FAM21A
|
family with sequence similarity 21, member C
family with sequence similarity 21, member A
|
|
chr17_-_7078399
|
2.223
|
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr2_+_219751145
|
2.222
|
NM_024293
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr12_-_108918570
|
2.210
|
NM_001135213
NM_001135214
NM_014776
NM_057169
NM_057170
NM_139201
|
GIT2
|
G protein-coupled receptor kinase interacting ArfGAP 2
|
|
chr6_+_123152044
|
2.204
|
|
SMPDL3A
|
sphingomyelin phosphodiesterase, acid-like 3A
|
|
chrX_-_100759562
|
2.183
|
NM_001009584
NM_001184768
NM_019007
|
ARMCX6
|
armadillo repeat containing, X-linked 6
|
|
chr6_+_42126228
|
2.178
|
NM_138572
|
TAF8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa
|
|
chr4_-_77353929
|
2.173
|
|
SCARB2
|
scavenger receptor class B, member 2
|
|
chr5_+_172343429
|
2.152
|
|
ATP6V0E1
|
ATPase, H+ transporting, lysosomal 9kDa, V0 subunit e1
|
|
chr2_+_241903395
|
2.150
|
NM_001008491
NM_006155
|
SEPT2
|
septin 2
|
|
chr3_-_79899748
|
2.139
|
NM_002941
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr2_+_231771623
|
2.135
|
|
ARMC9
|
armadillo repeat containing 9
|
|
chr14_+_61231795
|
2.118
|
NM_001530
NM_181054
|
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor)
|
|
chr10_+_121401061
|
2.118
|
|
BAG3
|
BCL2-associated athanogene 3
|