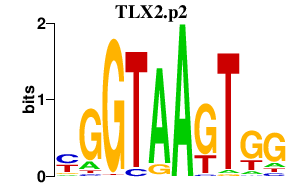
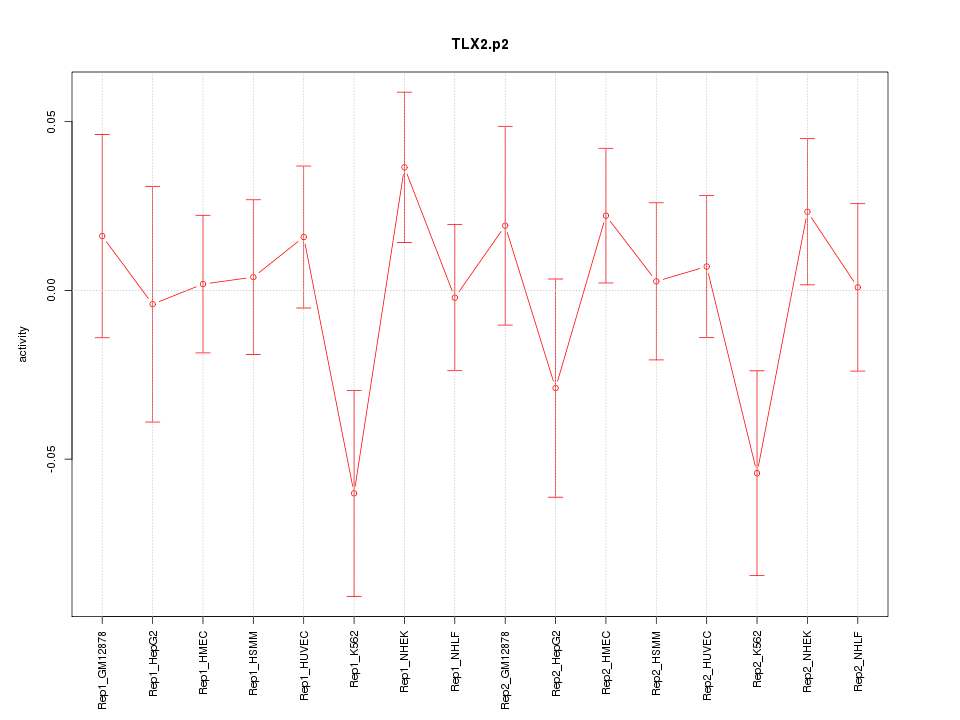
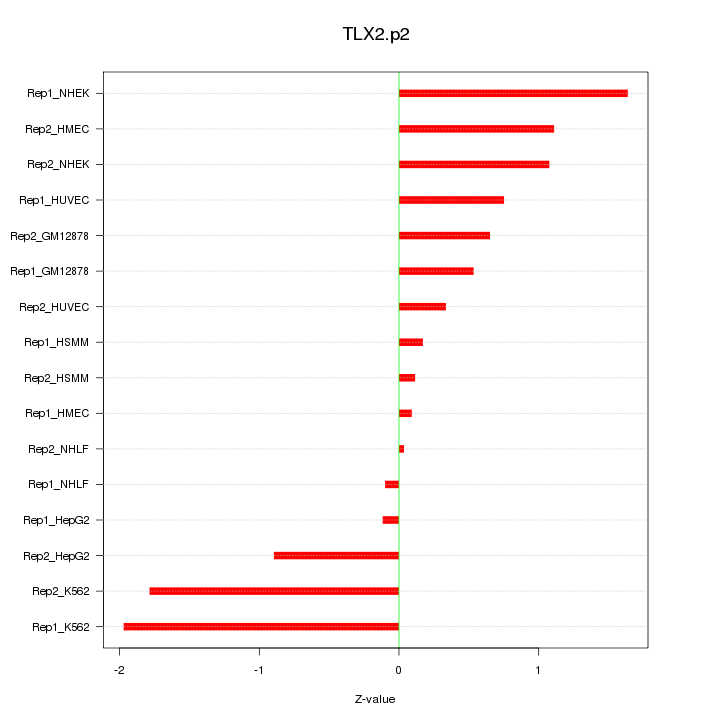
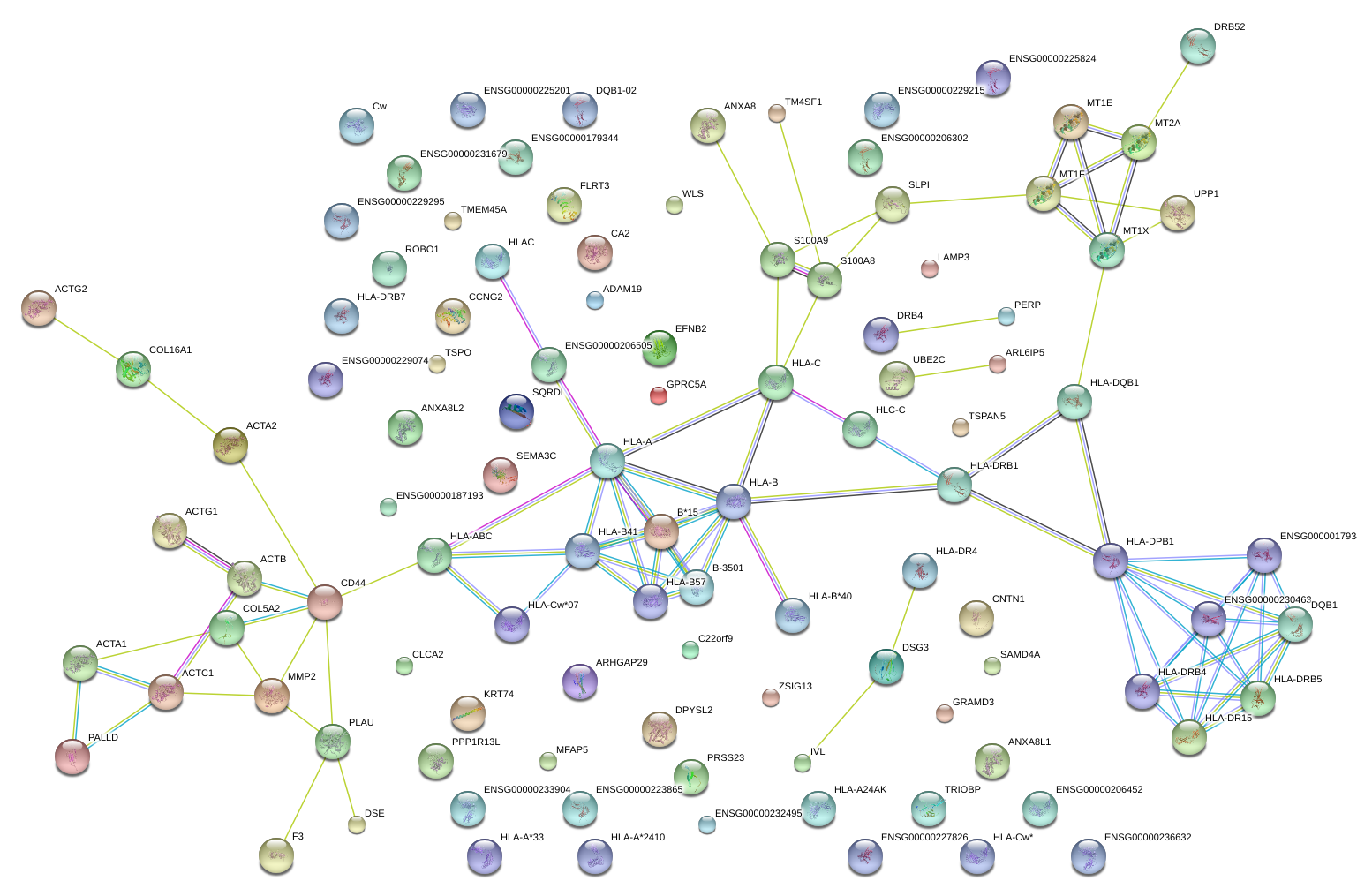
|
chr10_-_46593923
|
1.578
|
NM_001040084
NM_001098845
|
ANXA8L2
LOC653110
ANXA8
ANXA8L1
|
annexin A8-like 2
hypothetical LOC653110
annexin A8
annexin A8-like 1
|
|
chr10_+_47875229
|
1.508
|
NM_001040084
NM_001098845
|
ANXA8L2
ANXA8
ANXA8L1
|
annexin A8-like 2
annexin A8
annexin A8-like 1
|
|
chr10_+_47216925
|
1.452
|
NM_001630
|
ANXA8L2
ANXA8
|
annexin A8-like 2
annexin A8
|
|
chr4_+_169789327
|
1.321
|
NM_001166109
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr11_+_35117391
|
1.309
|
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr20_-_14266200
|
1.288
|
NM_013281
NM_198391
|
FLRT3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr6_+_116798791
|
1.234
|
NM_013352
|
DSE
|
dermatan sulfate epimerase
|
|
chr11_+_35117408
|
1.214
|
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr1_-_151630168
|
1.127
|
NM_002964
|
S100A8
|
S100 calcium binding protein A8
|
|
chr8_+_26491317
|
1.063
|
NM_001386
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr6_-_31432954
|
1.044
|
NM_005514
|
HLA-B
|
major histocompatibility complex, class I, B
|
|
chr1_-_68470585
|
0.940
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chr1_+_86662356
|
0.888
|
NM_006536
|
CLCA2
|
chloride channel accessory 2
|
|
chr19_-_50601406
|
0.867
|
NM_001142502
|
PPP1R13L
|
protein phosphatase 1, regulatory (inhibitor) subunit 13 like
|
|
chr11_+_86189138
|
0.861
|
NM_007173
|
PRSS23
|
protease, serine, 23
|
|
chr6_-_31432934
|
0.836
|
|
HLA-B
|
major histocompatibility complex, class I, B
|
|
chr1_+_151147646
|
0.796
|
NM_005547
|
IVL
|
involucrin
|
|
chr5_+_125786975
|
0.796
|
|
GRAMD3
|
GRAM domain containing 3
|
|
chr4_+_78297356
|
0.793
|
NM_004354
|
CCNG2
|
cyclin G2
|
|
chr3_-_78749633
|
0.792
|
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr22_+_36472262
|
0.781
|
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr7_+_48095464
|
0.760
|
|
UPP1
|
uridine phosphorylase 1
|
|
chr1_-_94779727
|
0.740
|
NM_001178096
NM_001993
|
F3
|
coagulation factor III (thromboplastin, tissue factor)
|
|
chr3_-_184363157
|
0.725
|
NM_014398
|
LAMP3
|
lysosomal-associated membrane protein 3
|
|
chr20_-_43316597
|
0.724
|
NM_003064
|
SLPI
|
secretory leukocyte peptidase inhibitor
|
|
chr4_+_78297418
|
0.718
|
|
CCNG2
|
cyclin G2
|
|
chr12_-_8706533
|
0.693
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr12_-_51253875
|
0.693
|
NM_175053
|
KRT74
|
keratin 74
|
|
chr16_+_54070302
|
0.689
|
NM_004530
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr3_-_150578023
|
0.663
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr16_+_55199978
|
0.661
|
NM_005953
|
MT2A
|
metallothionein 2A
|
|
chr1_+_151596953
|
0.657
|
NM_002965
|
S100A9
|
S100 calcium binding protein A9
|
|
chr10_-_90702490
|
0.645
|
NM_001613
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr22_-_44015424
|
0.643
|
|
KIAA0930
|
KIAA0930
|
|
chr20_+_43875078
|
0.640
|
NM_181801
|
UBE2C
|
ubiquitin-conjugating enzyme E2C
|
|
chr3_+_69216747
|
0.638
|
NM_006407
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5
|
|
chr22_+_36472176
|
0.620
|
NM_007032
NM_138632
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr12_+_12935759
|
0.618
|
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr7_-_80386258
|
0.614
|
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C
|
|
chr22_+_41877470
|
0.614
|
NM_000714
NM_007311
|
TSPO
|
translocator protein (18kDa)
|
|
chr13_-_105985313
|
0.612
|
NM_004093
|
EFNB2
|
ephrin-B2
|
|
chr10_+_75343352
|
0.611
|
|
PLAU
|
plasminogen activator, urokinase
|
|
chr1_-_94475840
|
0.610
|
NM_004815
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr12_+_12935619
|
0.610
|
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr22_+_41877502
|
0.609
|
|
TSPO
|
translocator protein (18kDa)
|
|
chr5_-_156935317
|
0.605
|
NM_033274
|
ADAM19
|
ADAM metallopeptidase domain 19
|
|
chr14_+_54104383
|
0.604
|
NM_001161576
NM_015589
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr1_-_94475504
|
0.597
|
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr4_-_99798563
|
0.591
|
NM_005723
|
TSPAN5
|
tetraspanin 5
|
|
chr7_+_48095263
|
0.580
|
NM_003364
|
UPP1
|
uridine phosphorylase 1
|
|
chr6_-_138470160
|
0.573
|
NM_022121
|
PERP
|
PERP, TP53 apoptosis effector
|
|
chr18_+_27281729
|
0.573
|
NM_001944
|
DSG3
|
desmoglein 3
|
|
chr2_-_189752575
|
0.565
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr8_+_86563382
|
0.564
|
NM_000067
|
CA2
|
carbonic anhydrase II
|
|
chr3_+_101694244
|
0.555
|
|
TMEM45A
|
transmembrane protein 45A
|
|
chr1_-_31942206
|
0.554
|
NM_001856
|
COL16A1
|
collagen, type XVI, alpha 1
|
|
chr15_+_43714500
|
0.540
|
NM_021199
|
SQRDL
|
sulfide quinone reductase-like (yeast)
|
|
chr6_-_32665409
|
0.532
|
NM_002124
|
HLA-DRB1
HLA-DRB4
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 1
major histocompatibility complex, class II, DR beta 4
major histocompatibility complex, class II, DR beta 5
|
|
chr12_+_15955549
|
0.529
|
|
DERA
|
deoxyribose-phosphate aldolase (putative)
|
|
chr13_+_109936080
|
0.519
|
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr6_-_31432877
|
0.517
|
|
HLA-B
|
major histocompatibility complex, class I, B
|
|
chrX_+_48253007
|
0.517
|
NM_203474
NM_203475
|
PORCN
|
porcupine homolog (Drosophila)
|
|
chr1_+_150753601
|
0.516
|
NM_019060
|
CRCT1
|
cysteine-rich C-terminal 1
|
|
chr11_+_59979857
|
0.515
|
NM_021950
NM_152866
|
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1
|
|
chr21_-_26175068
|
0.512
|
|
APP
|
amyloid beta (A4) precursor protein
|
|
chr4_-_35922288
|
0.511
|
NM_015230
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2
|
|
chr4_+_166468266
|
0.508
|
NM_001017369
NM_006745
|
SC4MOL
|
sterol-C4-methyl oxidase-like
|
|
chr9_+_123123364
|
0.508
|
|
GSN
|
gelsolin
|
|
chr10_-_103805858
|
0.502
|
NM_024541
|
C10orf76
|
chromosome 10 open reading frame 76
|
|
chr10_+_75340867
|
0.502
|
NM_001145031
NM_002658
|
PLAU
|
plasminogen activator, urokinase
|
|
chr1_-_31942142
|
0.494
|
|
COL16A1
|
collagen, type XVI, alpha 1
|
|
chr19_+_16296632
|
0.492
|
NM_016270
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr12_+_46785922
|
0.491
|
NM_001166686
|
PFKM
|
phosphofructokinase, muscle
|
|
chr5_-_150517499
|
0.484
|
|
ANXA6
|
annexin A6
|
|
chr10_+_3141596
|
0.483
|
|
PFKP
|
phosphofructokinase, platelet
|
|
chr7_-_120285364
|
0.476
|
NM_012338
|
TSPAN12
|
tetraspanin 12
|
|
chr18_-_59185330
|
0.471
|
NM_002035
|
KDSR
|
3-ketodihydrosphingosine reductase
|
|
chr20_+_56318415
|
0.463
|
|
RAB22A
|
RAB22A, member RAS oncogene family
|
|
chr2_-_20288108
|
0.461
|
NM_002997
|
SDC1
|
syndecan 1
|
|
chr4_+_38545741
|
0.455
|
NM_138389
|
FAM114A1
|
family with sequence similarity 114, member A1
|
|
chr12_+_15955372
|
0.454
|
NM_015954
|
DERA
|
deoxyribose-phosphate aldolase (putative)
|
|
chr6_+_32713160
|
0.452
|
NM_002122
|
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1
|
|
chr3_+_101720753
|
0.451
|
|
TMEM45A
|
transmembrane protein 45A
|
|
chrX_-_10761729
|
0.449
|
NM_033290
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr6_+_76515690
|
0.446
|
|
MYO6
|
myosin VI
|
|
chr22_+_29808845
|
0.446
|
|
SMTN
|
smoothelin
|
|
chr20_+_56318245
|
0.444
|
|
RAB22A
|
RAB22A, member RAS oncogene family
|
|
chr5_-_150517528
|
0.444
|
NM_001155
|
ANXA6
|
annexin A6
|
|
chr3_+_48482625
|
0.443
|
|
TREX1
|
three prime repair exonuclease 1
|
|
chr6_-_32742265
|
0.441
|
NM_002123
|
HLA-DQB1
|
major histocompatibility complex, class II, DQ beta 1
|
|
chr20_+_44180296
|
0.439
|
NM_001250
NM_152854
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5
|
|
chr6_-_112033752
|
0.435
|
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr4_-_90977149
|
0.434
|
NM_001146054
NM_007308
NM_000345
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chr22_-_44015248
|
0.430
|
NM_001009880
|
KIAA0930
|
KIAA0930
|
|
chr12_+_74160779
|
0.430
|
NM_006851
|
GLIPR1
|
GLI pathogenesis-related 1
|
|
chr7_-_87687276
|
0.429
|
|
SRI
|
sorcin
|
|
chr6_-_112186977
|
0.427
|
NM_153048
|
FYN
|
FYN oncogene related to SRC, FGR, YES
|
|
chr17_-_1037312
|
0.425
|
NM_001159746
|
ABR
|
active BCR-related gene
|
|
chr14_+_64240902
|
0.423
|
NM_015549
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3
|
|
chr12_-_94135154
|
0.422
|
NM_018351
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6
|
|
chr7_-_120285245
|
0.422
|
|
TSPAN12
|
tetraspanin 12
|
|
chr11_+_66827563
|
0.421
|
|
SSH3
|
slingshot homolog 3 (Drosophila)
|
|
chr16_+_31390567
|
0.421
|
NM_001042454
NM_001164719
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr3_+_101694098
|
0.419
|
NM_018004
|
TMEM45A
|
transmembrane protein 45A
|
|
chr20_+_33223434
|
0.418
|
NM_006404
|
PROCR
|
protein C receptor, endothelial
|
|
chr16_-_10581944
|
0.414
|
NM_001424
|
EMP2
|
epithelial membrane protein 2
|
|
chr10_+_63803921
|
0.411
|
NM_014951
NM_199450
NM_199451
|
ZNF365
|
zinc finger protein 365
|
|
chr5_+_96023926
|
0.410
|
|
CAST
|
calpastatin
|
|
chr6_+_76515608
|
0.410
|
NM_004999
|
MYO6
|
myosin VI
|
|
chr14_+_64041257
|
0.408
|
|
ZBTB1
|
zinc finger and BTB domain containing 1
|
|
chr3_-_55496607
|
0.404
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr17_-_3518568
|
0.402
|
|
TAX1BP3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3
|
|
chr11_+_66827494
|
0.401
|
NM_017857
|
SSH3
|
slingshot homolog 3 (Drosophila)
|
|
chr5_+_96023479
|
0.400
|
NM_001190442
|
CAST
|
calpastatin
|
|
chr15_-_40052041
|
0.398
|
NM_139265
|
EHD4
|
EH-domain containing 4
|
|
chr3_+_48482213
|
0.398
|
NM_016381
NM_033629
|
TREX1
|
three prime repair exonuclease 1
|
|
chr14_-_105308531
|
0.398
|
|
|
|
|
chr3_+_189425886
|
0.397
|
NM_001167671
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr1_+_167342493
|
0.397
|
NM_001677
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr17_-_3518628
|
0.396
|
|
TAX1BP3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3
|
|
chr22_+_36472201
|
0.395
|
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr3_-_45162865
|
0.393
|
|
CDCP1
|
CUB domain containing protein 1
|
|
chr6_+_32515596
|
0.390
|
NM_019111
|
HLA-DRA
|
major histocompatibility complex, class II, DR alpha
|
|
chr3_-_45162847
|
0.389
|
|
CDCP1
|
CUB domain containing protein 1
|
|
chr15_-_40051979
|
0.387
|
|
EHD4
|
EH-domain containing 4
|
|
chr9_+_115382990
|
0.386
|
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr5_+_96023627
|
0.386
|
NM_001042440
|
CAST
|
calpastatin
|
|
chr7_-_87687302
|
0.383
|
NM_003130
|
SRI
|
sorcin
|
|
chr1_+_154297588
|
0.379
|
NM_020387
|
RAB25
|
RAB25, member RAS oncogene family
|
|
chr10_-_116408046
|
0.373
|
NM_002313
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr7_-_150960086
|
0.373
|
NM_024429
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit
|
|
chr14_-_52487022
|
0.373
|
|
FERMT2
|
fermitin family member 2
|
|
chr3_-_180272291
|
0.372
|
NM_022470
NM_152240
|
ZMAT3
|
zinc finger, matrin-type 3
|
|
chr11_+_66580861
|
0.372
|
NM_014578
|
RHOD
|
ras homolog gene family, member D
|
|
chr1_+_163867057
|
0.370
|
NM_004528
|
MGST3
|
microsomal glutathione S-transferase 3
|
|
chr6_-_32605898
|
0.370
|
NM_002125
|
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 5
|
|
chr10_+_123744131
|
0.368
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr3_-_109292444
|
0.367
|
NM_001777
NM_198793
|
CD47
|
CD47 molecule
|
|
chr20_+_56318155
|
0.366
|
NM_020673
|
RAB22A
|
RAB22A, member RAS oncogene family
|
|
chr11_+_19328846
|
0.366
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chrX_-_10605726
|
0.363
|
NM_001098624
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr3_-_122951200
|
0.361
|
NM_004487
|
GOLGB1
|
golgin B1
|
|
chr5_-_150517454
|
0.360
|
|
ANXA6
|
annexin A6
|
|
chr14_-_105796665
|
0.360
|
|
IGHV4-31
IGHV3-23
IGHA1
IGHG1
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy variable 3-23
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant mu
|
|
chr6_-_33028790
|
0.358
|
NM_006120
|
HLA-DMA
|
major histocompatibility complex, class II, DM alpha
|
|
chr10_+_75340921
|
0.355
|
|
PLAU
|
plasminogen activator, urokinase
|
|
chr1_-_151333624
|
0.355
|
NM_001024209
|
SPRR2E
|
small proline-rich protein 2E
|
|
chr14_-_22358752
|
0.353
|
NM_001126106
|
SLC7A7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7
|
|
chr6_-_16869579
|
0.347
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chr2_+_102345528
|
0.345
|
NM_003855
|
IL18R1
|
interleukin 18 receptor 1
|
|
chr20_+_42537927
|
0.344
|
NM_001039199
NM_024331
|
TTPAL
|
tocopherol (alpha) transfer protein-like
|
|
chr6_-_109883639
|
0.343
|
|
MICAL1
|
microtubule associated monoxygenase, calponin and LIM domain containing 1
|
|
chr5_+_172343415
|
0.343
|
|
ATP6V0E1
|
ATPase, H+ transporting, lysosomal 9kDa, V0 subunit e1
|
|
chr8_-_42184350
|
0.342
|
NM_000930
NM_033011
|
PLAT
|
plasminogen activator, tissue
|
|
chr14_-_54025414
|
0.340
|
|
GMFB
|
glia maturation factor, beta
|
|
chr12_-_8706746
|
0.340
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr13_-_37070862
|
0.340
|
NM_001135934
NM_001135935
NM_001135936
NM_006475
|
POSTN
|
periostin, osteoblast specific factor
|
|
chr19_+_48772791
|
0.339
|
NM_001193621
|
LOC390940
|
hypothetical protein LOC390940
|
|
chr1_-_85816520
|
0.339
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr13_-_32658215
|
0.338
|
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr2_-_110230642
|
0.336
|
|
MALL
|
mal, T-cell differentiation protein-like
|
|
chr19_-_11549508
|
0.335
|
NM_001611
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr5_+_36642213
|
0.335
|
NM_001166696
NM_004172
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr3_+_123256902
|
0.334
|
NM_175862
|
CD86
|
CD86 molecule
|
|
chr3_-_45162901
|
0.332
|
NM_022842
NM_178181
|
CDCP1
|
CUB domain containing protein 1
|
|
chr3_-_180272229
|
0.332
|
|
ZMAT3
|
zinc finger, matrin-type 3
|
|
chr20_-_1321702
|
0.332
|
|
FKBP1A-SDCBP2
FKBP1A
|
FKBP1A-SDCBP2 read-through transcript
FK506 binding protein 1A, 12kDa
|
|
chr12_-_55791443
|
0.332
|
NM_001178078
NM_001178080
NM_001178081
NM_003153
|
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced
|
|
chr14_-_105181939
|
0.330
|
|
IGHG2
|
immunoglobulin heavy constant gamma 2 (G2m marker)
|
|
chr10_+_123862430
|
0.330
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr14_-_54025464
|
0.328
|
NM_004124
|
GMFB
|
glia maturation factor, beta
|
|
chr15_-_57452361
|
0.326
|
NM_004998
|
MYO1E
|
myosin IE
|
|
chr16_-_11588257
|
0.326
|
NM_001136472
NM_001136473
|
LITAF
|
lipopolysaccharide-induced TNF factor
|
|
chr3_-_152517204
|
0.326
|
NM_023915
|
GPR87
|
G protein-coupled receptor 87
|
|
chr17_-_3518654
|
0.325
|
NM_014604
|
TAX1BP3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3
|
|
chr1_-_85515174
|
0.324
|
NM_003921
|
BCL10
|
B-cell CLL/lymphoma 10
|
|
chr2_+_64535114
|
0.324
|
|
HSPC159
|
galectin-related protein
|
|
chr16_-_66827449
|
0.323
|
NM_024939
|
ESRP2
|
epithelial splicing regulatory protein 2
|
|
chrX_+_51562894
|
0.323
|
NM_001005332
|
MAGED1
|
melanoma antigen family D, 1
|
|
chr12_+_52618815
|
0.323
|
NM_017410
|
HOXC13
|
homeobox C13
|
|
chr12_-_52960168
|
0.319
|
NM_012117
|
CBX5
|
chromobox homolog 5
|
|
chr2_+_64534774
|
0.319
|
NM_014181
|
HSPC159
|
galectin-related protein
|
|
chr12_-_8706637
|
0.319
|
NM_003480
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr19_-_8548232
|
0.319
|
|
MYO1F
|
myosin IF
|
|
chr19_+_627346
|
0.318
|
NM_005860
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein)
|
|
chr20_-_1321571
|
0.318
|
|
FKBP1A
|
FK506 binding protein 1A, 12kDa
|
|
chr22_+_40559070
|
0.317
|
|
SREBF2
|
sterol regulatory element binding transcription factor 2
|
|
chr6_-_109883635
|
0.316
|
|
MICAL1
|
microtubule associated monoxygenase, calponin and LIM domain containing 1
|
|
chr11_+_13255913
|
0.316
|
|
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like
|
|
chr15_-_57452345
|
0.316
|
|
MYO1E
|
myosin IE
|
|
chr1_-_85703198
|
0.315
|
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr2_-_85969527
|
0.315
|
NM_003896
|
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5
|
|
chr6_-_30289092
|
0.313
|
NM_003449
|
TRIM26
|
tripartite motif containing 26
|
|
chr5_+_172343429
|
0.313
|
|
ATP6V0E1
|
ATPase, H+ transporting, lysosomal 9kDa, V0 subunit e1
|
|
chr7_+_90063543
|
0.312
|
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr6_-_31658044
|
0.312
|
NM_002341
NM_009588
|
LTB
|
lymphotoxin beta (TNF superfamily, member 3)
|
|
chr13_+_113023938
|
0.307
|
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chrX_-_70067650
|
0.305
|
NM_001048164
NM_032803
|
SLC7A3
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3
|
|
chr1_-_25164061
|
0.304
|
NM_001031680
|
RUNX3
|
runt-related transcription factor 3
|
|
chr11_+_13255948
|
0.304
|
|
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like
|