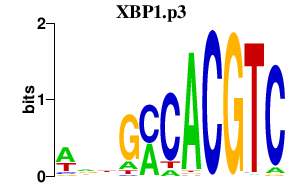
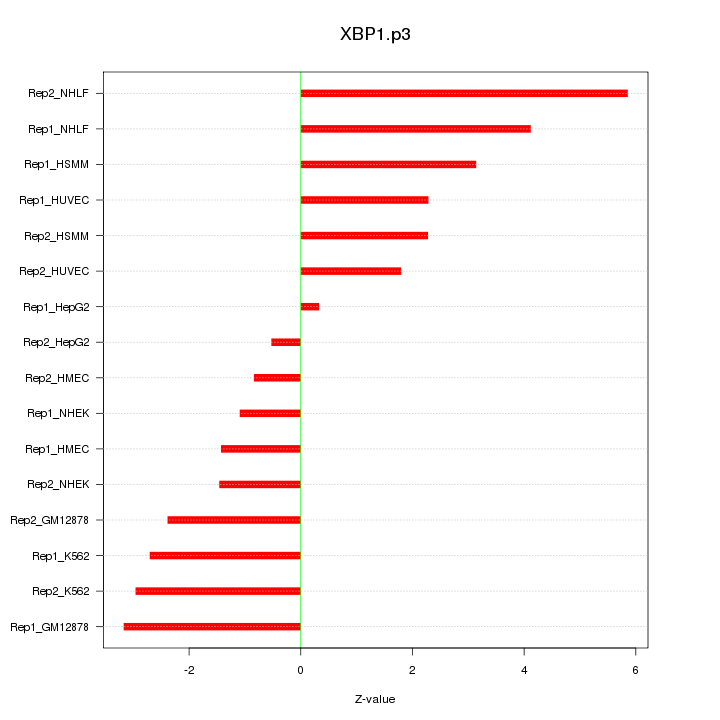
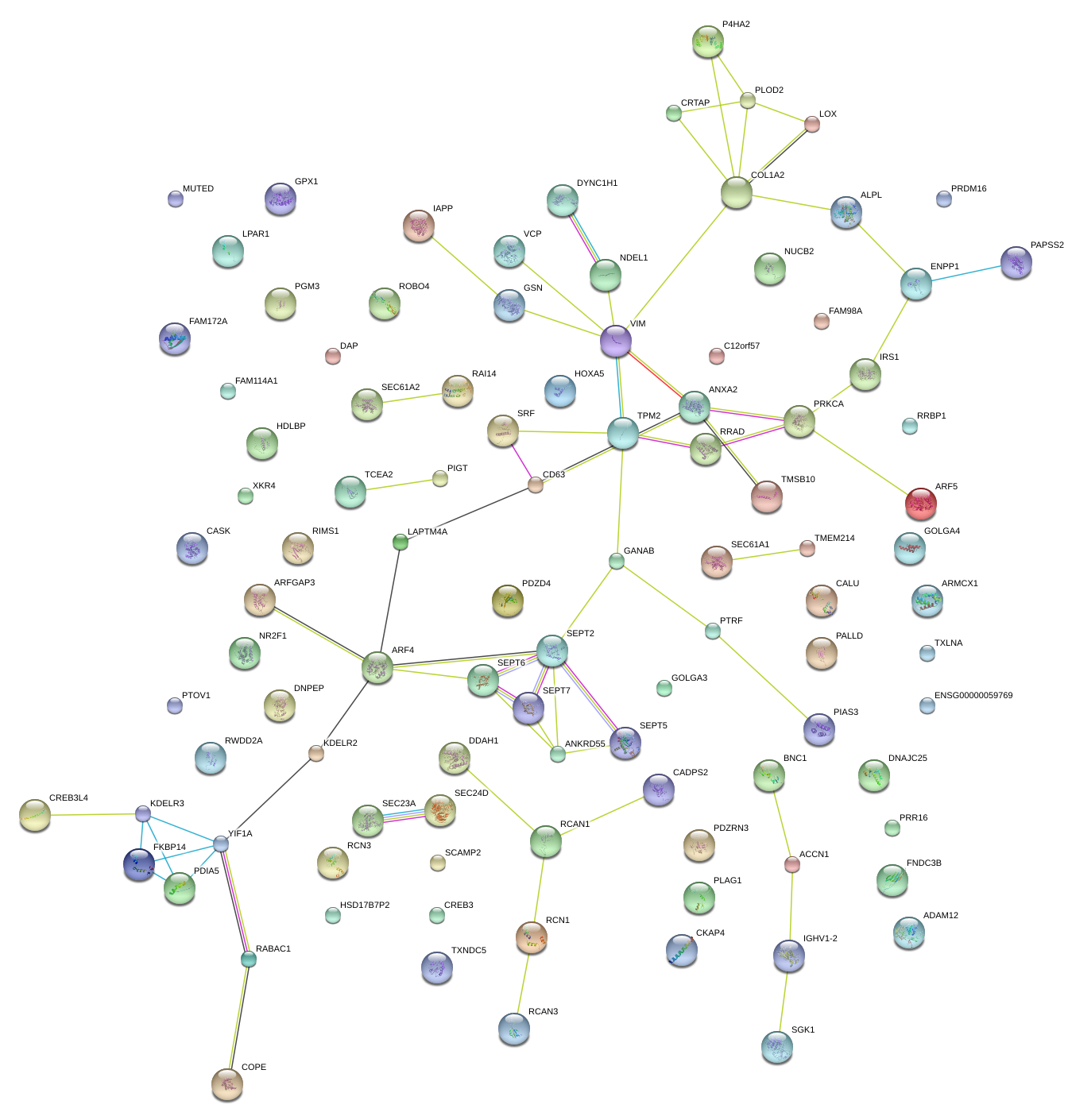
|
chr7_+_93861808
|
16.260
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr19_+_54723044
|
8.582
|
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr5_-_121441910
|
8.466
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr10_+_89409332
|
7.327
|
NM_001015880
NM_004670
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2
|
|
chr5_-_121441805
|
6.158
|
|
LOX
|
lysyl oxidase
|
|
chr12_-_105165805
|
5.756
|
NM_006825
|
CKAP4
|
cytoskeleton-associated protein 4
|
|
chr4_+_38545741
|
5.717
|
NM_138389
|
FAM114A1
|
family with sequence similarity 114, member A1
|
|
chr5_-_121441866
|
5.658
|
|
LOX
|
lysyl oxidase
|
|
chr12_+_6923441
|
4.627
|
NM_138425
|
C12orf57
|
chromosome 12 open reading frame 57
|
|
chr3_-_57558193
|
4.582
|
NM_001660
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr7_+_128166581
|
4.538
|
NM_001130674
NM_001219
|
CALU
|
calumenin
|
|
chr22_+_37193961
|
4.453
|
NM_006855
NM_016657
|
KDELR3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3
|
|
chr19_+_54722686
|
4.235
|
NM_020650
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr7_-_6490214
|
4.091
|
NM_001100603
NM_006854
|
KDELR2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2
|
|
chr5_+_119827903
|
3.837
|
NM_016644
|
PRR16
|
proline rich 16
|
|
chr3_-_57558130
|
3.816
|
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr3_-_57558120
|
3.740
|
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr3_-_57557906
|
3.577
|
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr3_+_173240983
|
3.523
|
NM_001135095
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr7_+_128166664
|
3.333
|
|
CALU
|
calumenin
|
|
chr7_+_128166631
|
3.330
|
|
CALU
|
calumenin
|
|
chr7_+_128166636
|
3.270
|
|
CALU
|
calumenin
|
|
chr7_+_128166688
|
3.011
|
|
CALU
|
calumenin
|
|
chr11_+_32068990
|
2.839
|
NM_002901
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain
|
|
chr5_+_119827797
|
2.799
|
|
PRR16
|
proline rich 16
|
|
chr5_-_93472993
|
2.783
|
NM_001163417
NM_001163418
NM_032042
|
FAM172A
|
family with sequence similarity 172, member A
|
|
chr20_-_17610828
|
2.752
|
NM_001042576
NM_004587
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr11_+_32069183
|
2.605
|
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain
|
|
chr7_-_27149750
|
2.600
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr4_-_119976631
|
2.575
|
|
SEC24D
|
SEC24 family, member D (S. cerevisiae)
|
|
chr2_-_20115269
|
2.459
|
NM_014713
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha
|
|
chr6_-_83959640
|
2.370
|
NM_015599
|
PGM3
|
phosphoglucomutase 3
|
|
chr14_-_38642165
|
2.245
|
NM_006364
|
SEC23A
|
Sec23 homolog A (S. cerevisiae)
|
|
chr17_-_37828643
|
2.194
|
NM_012232
|
PTRF
|
polymerase I and transcript release factor
|
|
chr10_-_128067062
|
2.174
|
NM_003474
NM_021641
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr2_+_241903395
|
2.156
|
NM_001008491
NM_006155
|
SEPT2
|
septin 2
|
|
chr7_-_6490375
|
2.138
|
|
KDELR2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2
|
|
chr7_+_93862137
|
2.110
|
|
COL1A2
PLAG1
|
collagen, type I, alpha 2
pleiomorphic adenoma gene 1
|
|
chr6_-_134538526
|
2.094
|
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr7_-_30032649
|
2.045
|
NM_017946
|
FKBP14
|
FK506 binding protein 14, 22 kDa
|
|
chr5_-_131591380
|
2.034
|
NM_001017974
NM_001142598
NM_001142599
|
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II
|
|
chr12_-_54409710
|
2.027
|
|
CD63
|
CD63 molecule
|
|
chr4_-_119976736
|
2.018
|
NM_014822
|
SEC24D
|
SEC24 family, member D (S. cerevisiae)
|
|
chr2_-_20114881
|
1.992
|
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha
|
|
chr12_-_131915270
|
1.958
|
NM_005895
|
GOLGA3
|
golgin A3
|
|
chr20_-_17610704
|
1.952
|
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr8_+_56177567
|
1.933
|
NM_052898
|
XKR4
|
XK, Kell blood group complex subunit-related family, member 4
|
|
chr15_-_58477407
|
1.924
|
NM_001002857
NM_001002858
NM_001136015
NM_004039
|
ANXA2
|
annexin A2
|
|
chr6_+_132170888
|
1.904
|
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chr20_+_43478120
|
1.904
|
NM_001184728
NM_001184729
NM_001184730
NM_015937
|
PIGT
|
phosphatidylinositol glycan anchor biosynthesis, class T
|
|
chr3_-_49370755
|
1.826
|
NM_000581
NM_201397
|
GPX1
|
glutathione peroxidase 1
|
|
chr3_+_129253816
|
1.779
|
NM_013336
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae)
|
|
chr1_-_85703198
|
1.770
|
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr3_+_33130451
|
1.767
|
NM_006371
|
CRTAP
|
cartilage associated protein
|
|
chr14_-_38642029
|
1.658
|
|
SEC23A
|
Sec23 homolog A (S. cerevisiae)
|
|
chr3_+_129253977
|
1.631
|
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae)
|
|
chr2_-_20114761
|
1.590
|
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha
|
|
chr9_+_35722629
|
1.588
|
|
CREB3
|
cAMP responsive element binding protein 3
|
|
chr6_+_83959750
|
1.550
|
NM_033411
|
RWDD2A
|
RWD domain containing 2A
|
|
chr3_+_33130563
|
1.545
|
|
CRTAP
|
cartilage associated protein
|
|
chr14_+_101500520
|
1.538
|
NM_001376
|
DYNC1H1
|
dynein, cytoplasmic 1, heavy chain 1
|
|
chr4_+_169789327
|
1.506
|
NM_001166109
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr9_-_35679902
|
1.480
|
NM_003289
NM_213674
|
TPM2
|
tropomyosin 2 (beta)
|
|
chr9_+_35722207
|
1.455
|
NM_006368
|
CREB3
|
cAMP responsive element binding protein 3
|
|
chr3_-_73756668
|
1.425
|
NM_015009
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr6_-_7855125
|
1.421
|
NM_001145549
|
TXNDC5
|
thioredoxin domain containing 5 (endoplasmic reticulum)
|
|
chrX_+_100692071
|
1.415
|
NM_016608
|
ARMCX1
|
armadillo repeat containing, X-linked 1
|
|
chr9_+_35722490
|
1.411
|
|
CREB3
|
cAMP responsive element binding protein 3
|
|
chr6_+_132170847
|
1.409
|
NM_006208
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chrX_-_41667174
|
1.396
|
NM_001126054
NM_001126055
NM_003688
|
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family)
|
|
chr2_-_241903657
|
1.390
|
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr14_+_101500650
|
1.377
|
|
DYNC1H1
|
dynein, cytoplasmic 1, heavy chain 1
|
|
chr6_+_72949092
|
1.306
|
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr6_-_7855715
|
1.296
|
NM_030810
|
TXNDC5
|
thioredoxin domain containing 5 (endoplasmic reticulum)
|
|
chr2_+_84986510
|
1.286
|
|
TMSB10
|
thymosin beta 10
|
|
chr9_-_112840064
|
1.276
|
NM_001401
NM_057159
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr9_+_113433364
|
1.264
|
NM_001015882
NM_004125
|
DNAJC25
DNAJC25-GNG10
|
DnaJ (Hsp40) homolog, subfamily C , member 25
DNAJC25-GNG10 readthrough
|
|
chr11_+_17254854
|
1.248
|
NM_005013
|
NUCB2
|
nucleobindin 2
|
|
chr6_+_1336077
|
1.244
|
|
|
|
|
chr3_+_124268545
|
1.244
|
NM_006810
|
PDIA5
|
protein disulfide isomerase family A, member 5
|
|
chr5_+_92946348
|
1.236
|
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr11_-_65813031
|
1.211
|
|
YIF1A
|
Yip1 interacting factor homolog A (S. cerevisiae)
|
|
chr5_+_34692124
|
1.152
|
NM_001145522
NM_015577
|
RAI14
|
retinoic acid induced 14
|
|
chr15_-_72952648
|
1.150
|
NM_005697
|
SCAMP2
|
secretory carrier membrane protein 2
|
|
chr16_-_65516919
|
1.147
|
NM_001128850
NM_004165
|
RRAD
|
Ras-related associated with diabetes
|
|
chr10_+_17312654
|
1.146
|
|
VIM
|
vimentin
|
|
chr10_-_128067001
|
1.146
|
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr2_+_27109249
|
1.103
|
NM_001083590
NM_017727
|
TMEM214
|
transmembrane protein 214
|
|
chr3_+_37259684
|
1.088
|
NM_001172713
NM_002078
|
GOLGA4
|
golgin A4
|
|
chr17_+_61729306
|
1.022
|
NM_002737
|
PRKCA
|
protein kinase C, alpha
|
|
chr2_-_33677798
|
1.013
|
NM_015475
|
FAM98A
|
family with sequence similarity 98, member A
|
|
chr19_+_55045754
|
1.009
|
|
PTOV1
|
prostate tumor overexpressed 1
|
|
chr1_+_144287368
|
1.009
|
|
PIAS3
|
protein inhibitor of activated STAT, 3
|
|
chr2_+_84986273
|
0.966
|
NM_021103
|
TMSB10
|
thymosin beta 10
|
|
chr9_+_123003549
|
0.958
|
|
GSN
|
gelsolin
|
|
chr1_+_21708428
|
0.952
|
NM_000478
NM_001127501
NM_001177520
|
ALPL
|
alkaline phosphatase, liver/bone/kidney
|
|
chr2_+_27109341
|
0.948
|
|
TMEM214
|
transmembrane protein 214
|
|
chr17_+_8279860
|
0.943
|
NM_001025579
NM_030808
|
NDEL1
|
nudE nuclear distribution gene E homolog (A. nidulans)-like 1
|
|
chr19_-_47155340
|
0.928
|
NM_006423
|
RABAC1
|
Rab acceptor 1 (prenylated)
|
|
chr22_-_41583130
|
0.921
|
|
ARFGAP3
|
ADP-ribosylation factor GTPase activating protein 3
|
|
chr3_-_147361465
|
0.918
|
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr15_-_81744469
|
0.915
|
NM_001717
|
BNC1
|
basonuclin 1
|
|
chr1_+_32418234
|
0.903
|
|
TXLNA
|
taxilin alpha
|
|
chr7_-_122313646
|
0.897
|
|
CADPS2
|
Ca++-dependent secretion activator 2
|
|
chr20_+_62164453
|
0.895
|
NM_003195
|
TCEA2
|
transcription elongation factor A (SII), 2
|
|
chr11_-_65813183
|
0.873
|
NM_020470
|
YIF1A
|
Yip1 interacting factor homolog A (S. cerevisiae)
|
|
chr2_-_227371698
|
0.873
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr9_-_35062545
|
0.872
|
|
VCP
|
valosin containing protein
|
|
chr1_+_144287344
|
0.857
|
NM_006099
|
PIAS3
|
protein inhibitor of activated STAT, 3
|
|
chr11_-_62170639
|
0.851
|
|
GANAB
|
glucosidase, alpha; neutral AB
|
|
chr19_-_18891197
|
0.850
|
NM_007263
NM_199442
NM_199444
|
COPE
|
coatomer protein complex, subunit epsilon
|
|
chr5_-_10814344
|
0.844
|
NM_004394
|
DAP
|
death-associated protein
|
|
chr2_+_176761552
|
0.839
|
NM_024501
|
HOXD1
|
homeobox D1
|
|
chr22_-_41583248
|
0.838
|
NM_001142293
NM_014570
|
ARFGAP3
|
ADP-ribosylation factor GTPase activating protein 3
|
|
chr5_-_9599157
|
0.834
|
NM_003966
|
SEMA5A
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A
|
|
chr3_-_49370682
|
0.821
|
|
GPX1
|
glutathione peroxidase 1
|
|
chr19_-_18891190
|
0.820
|
|
COPE
|
coatomer protein complex, subunit epsilon
|
|
chr11_+_117948301
|
0.814
|
NM_001142281
NM_001655
|
ARCN1
|
archain 1
|
|
chr1_+_52840630
|
0.813
|
NM_015696
|
GPX6
GPX7
|
glutathione peroxidase 6 (olfactory)
glutathione peroxidase 7
|
|
chr21_+_41461597
|
0.802
|
NM_012105
NM_138991
NM_138992
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chr11_+_117948350
|
0.802
|
|
ARCN1
|
archain 1
|
|
chr20_+_61054539
|
0.800
|
|
SLC17A9
|
solute carrier family 17, member 9
|
|
chr19_-_18891112
|
0.799
|
|
COPE
|
coatomer protein complex, subunit epsilon
|
|
chr2_-_99319197
|
0.788
|
NM_005783
|
TXNDC9
|
thioredoxin domain containing 9
|
|
chr20_+_25176698
|
0.784
|
NM_002862
|
PYGB
|
phosphorylase, glycogen; brain
|
|
chr9_-_35722175
|
0.770
|
|
TLN1
|
talin 1
|
|
chr11_-_62170672
|
0.764
|
NM_198334
NM_198335
|
GANAB
|
glucosidase, alpha; neutral AB
|
|
chr10_-_98336783
|
0.754
|
NM_020123
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr3_-_151171190
|
0.742
|
NM_002628
NM_053024
|
PFN2
|
profilin 2
|
|
chr6_-_134538762
|
0.736
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr14_+_99329198
|
0.732
|
NM_001008707
NM_004434
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr22_-_41583182
|
0.730
|
|
ARFGAP3
|
ADP-ribosylation factor GTPase activating protein 3
|
|
chr10_-_71600191
|
0.728
|
NM_001142648
NM_020150
|
SAR1A
|
SAR1 homolog A (S. cerevisiae)
|
|
chr19_-_18891178
|
0.723
|
|
COPE
|
coatomer protein complex, subunit epsilon
|
|
chr5_+_126881207
|
0.718
|
NM_130809
|
PRRC1
|
proline-rich coiled-coil 1
|
|
chr1_-_143643324
|
0.715
|
NM_001002811
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr19_-_18891196
|
0.699
|
|
COPE
|
coatomer protein complex, subunit epsilon
|
|
chr2_-_101134146
|
0.689
|
NM_001102426
|
TBC1D8
|
TBC1 domain family, member 8 (with GRAM domain)
|
|
chr19_-_18891188
|
0.678
|
|
COPE
|
coatomer protein complex, subunit epsilon
|
|
chr5_-_108773468
|
0.675
|
NM_014819
|
PJA2
|
praja ring finger 2
|
|
chr3_-_122951200
|
0.671
|
NM_004487
|
GOLGB1
|
golgin B1
|
|
chr19_+_977271
|
0.665
|
NM_004368
NM_201277
|
CNN2
|
calponin 2
|
|
chr6_-_131426014
|
0.661
|
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2
|
|
chr9_-_35722389
|
0.657
|
NM_006289
|
TLN1
|
talin 1
|
|
chr9_-_35722115
|
0.657
|
|
TLN1
|
talin 1
|
|
chr2_-_69467825
|
0.656
|
NM_002056
|
GFPT1
|
glutamine--fructose-6-phosphate transaminase 1
|
|
chr11_-_62170662
|
0.654
|
|
GANAB
|
glucosidase, alpha; neutral AB
|
|
chr2_+_202607554
|
0.645
|
NM_003507
|
FZD7
|
frizzled homolog 7 (Drosophila)
|
|
chr5_-_10814179
|
0.638
|
|
DAP
|
death-associated protein
|
|
chr5_-_10814341
|
0.635
|
|
DAP
|
death-associated protein
|
|
chr20_+_61054443
|
0.614
|
NM_022082
|
SLC17A9
|
solute carrier family 17, member 9
|
|
chr14_+_92330383
|
0.608
|
NM_005113
|
GOLGA5
|
golgin A5
|
|
chr19_-_18891205
|
0.602
|
|
COPE
|
coatomer protein complex, subunit epsilon
|
|
chrX_+_67635593
|
0.599
|
|
YIPF6
|
Yip1 domain family, member 6
|
|
chr14_+_73555802
|
0.598
|
NM_025057
|
C14orf45
|
chromosome 14 open reading frame 45
|
|
chr15_+_88610181
|
0.596
|
|
NGRN
|
neugrin, neurite outgrowth associated
|
|
chr10_-_98336584
|
0.593
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr9_-_35062596
|
0.590
|
|
VCP
|
valosin containing protein
|
|
chr3_-_188945921
|
0.589
|
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr19_+_977582
|
0.588
|
|
CNN2
|
calponin 2
|
|
chr19_-_18891195
|
0.588
|
|
COPE
|
coatomer protein complex, subunit epsilon
|
|
chr9_-_35062578
|
0.580
|
|
VCP
|
valosin containing protein
|
|
chrX_+_146941222
|
0.577
|
|
FTH1P8
|
ferritin, heavy polypeptide 1 pseudogene 8
|
|
chr1_+_110328850
|
0.577
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr2_+_183289205
|
0.573
|
NM_018981
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10
|
|
chr11_+_64608267
|
0.553
|
NM_006782
|
ZFPL1
|
zinc finger protein-like 1
|
|
chr19_-_47155277
|
0.553
|
|
RABAC1
|
Rab acceptor 1 (prenylated)
|
|
chr1_+_110328922
|
0.545
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr12_-_131915023
|
0.544
|
|
GOLGA3
|
golgin A3
|
|
chr5_+_34692239
|
0.543
|
|
RAI14
|
retinoic acid induced 14
|
|
chr16_+_55523526
|
0.538
|
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1
|
|
chr3_-_49736297
|
0.535
|
NM_013334
NM_021971
|
GMPPB
|
GDP-mannose pyrophosphorylase B
|
|
chr13_-_95503566
|
0.532
|
NM_020121
|
UGGT2
|
UDP-glucose glycoprotein glucosyltransferase 2
|
|
chr11_-_72986738
|
0.530
|
NM_015159
|
FAM168A
|
family with sequence similarity 168, member A
|
|
chr12_-_54408391
|
0.529
|
|
CD63
|
CD63 molecule
|
|
chr7_-_19123787
|
0.528
|
NM_000474
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr19_-_4621375
|
0.523
|
NM_019107
|
C19orf10
|
chromosome 19 open reading frame 10
|
|
chr12_+_50750013
|
0.520
|
NM_001098673
NM_021934
|
C12orf44
|
chromosome 12 open reading frame 44
|
|
chr2_-_110230927
|
0.515
|
|
MALL
|
mal, T-cell differentiation protein-like
|
|
chr9_-_35062643
|
0.514
|
NM_007126
|
VCP
|
valosin containing protein
|
|
chr19_-_4621323
|
0.508
|
|
C19orf10
|
chromosome 19 open reading frame 10
|
|
chr14_-_23734598
|
0.506
|
NM_001014842
NM_006405
|
TM9SF1
|
transmembrane 9 superfamily member 1
|
|
chr19_+_977627
|
0.503
|
|
CNN2
|
calponin 2
|
|
chr5_+_34692352
|
0.501
|
NM_001145520
|
RAI14
|
retinoic acid induced 14
|
|
chr14_+_49157150
|
0.493
|
NM_002408
|
MGAT2
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase
|
|
chr19_+_41298493
|
0.489
|
|
TBCB
|
tubulin folding cofactor B
|
|
chr4_+_25931282
|
0.482
|
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region
|
|
chr10_-_16899342
|
0.481
|
|
RSU1
|
Ras suppressor protein 1
|
|
chr6_+_116798791
|
0.475
|
NM_013352
|
DSE
|
dermatan sulfate epimerase
|
|
chr6_-_43592679
|
0.466
|
NM_015388
|
YIPF3
|
Yip1 domain family, member 3
|
|
chr15_+_41825941
|
0.464
|
|
PDIA3
|
protein disulfide isomerase family A, member 3
|
|
chr1_+_86943074
|
0.462
|
|
SH3GLB1
|
SH3-domain GRB2-like endophilin B1
|
|
chr3_-_107070400
|
0.451
|
NM_170662
|
CBLB
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence b
|
|
chr1_+_86942810
|
0.450
|
NM_016009
|
SH3GLB1
|
SH3-domain GRB2-like endophilin B1
|
|
chr19_+_977608
|
0.449
|
|
CNN2
|
calponin 2
|
|
chr3_+_130451119
|
0.432
|
NM_016128
|
COPG
|
coatomer protein complex, subunit gamma
|
|
chr4_+_8645421
|
0.430
|
|
CPZ
|
carboxypeptidase Z
|
|
chr1_+_110328941
|
0.426
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr1_+_32417909
|
0.425
|
NM_175852
|
TXLNA
|
taxilin alpha
|
|
chrX_-_15782859
|
0.421
|
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|