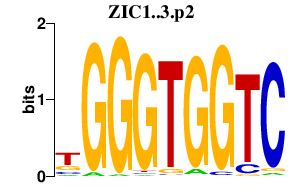
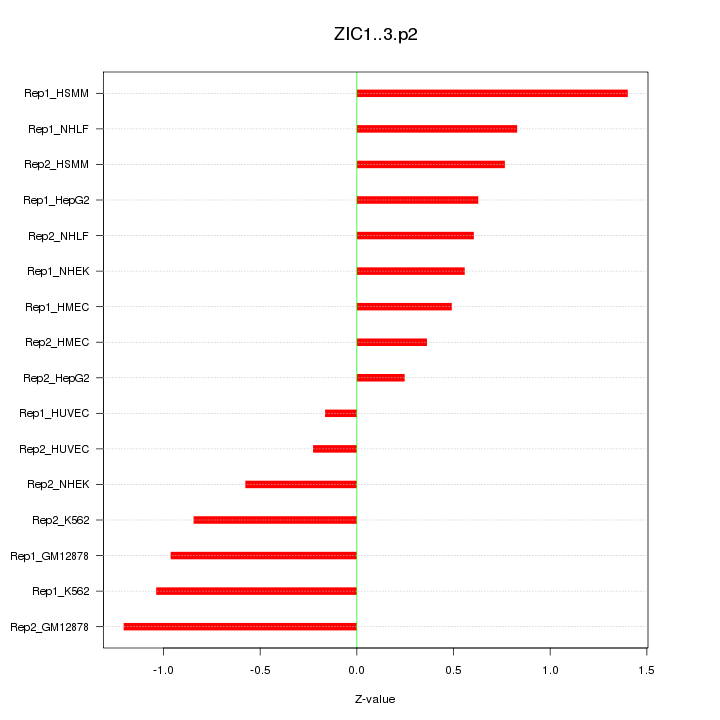
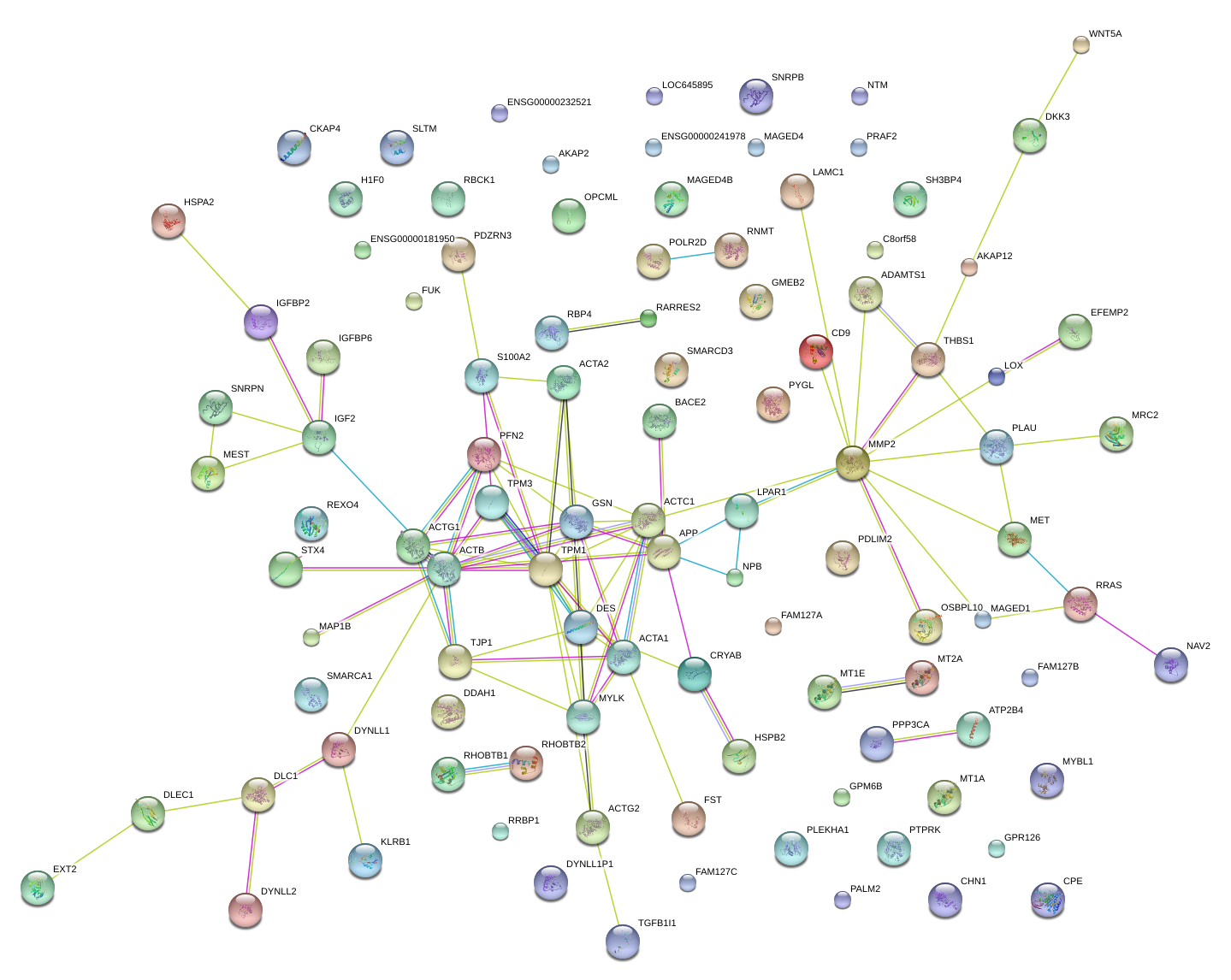
|
chr5_+_52812165
|
1.634
|
|
FST
|
follistatin
|
|
chr15_+_37660568
|
1.590
|
NM_003246
|
THBS1
|
thrombospondin 1
|
|
chr16_+_69060325
|
1.249
|
|
FUK
|
fucokinase
|
|
chr5_-_121440704
|
1.193
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr11_-_11987115
|
1.189
|
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr11_-_111287657
|
1.184
|
NM_001885
|
CRYAB
|
crystallin, alpha B
|
|
chr5_+_52812236
|
1.174
|
NM_006350
NM_013409
|
FST
|
follistatin
|
|
chr10_-_90702490
|
1.118
|
NM_001613
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr11_-_65396775
|
1.091
|
NM_016938
|
EFEMP2
|
EGF containing fibulin-like extracellular matrix protein 2
|
|
chr21_+_41461962
|
1.078
|
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chr11_-_11986704
|
0.993
|
NM_001018057
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr11_+_19691397
|
0.985
|
NM_145117
NM_182964
|
NAV2
|
neuron navigator 2
|
|
chr16_+_31390567
|
0.975
|
NM_001042454
NM_001164719
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr15_+_61127367
|
0.974
|
NM_001018008
|
TPM1
|
tropomyosin 1 (alpha)
|
|
chr11_-_11987198
|
0.974
|
NM_015881
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr10_+_75343352
|
0.955
|
|
PLAU
|
plasminogen activator, urokinase
|
|
chr3_-_73756668
|
0.948
|
NM_015009
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr11_+_111288669
|
0.938
|
NM_001541
|
HSPB2-C11ORF52
HSPB2
|
HSPB2-C11orf52 read-through transcript
heat shock 27kDa protein 2
|
|
chr11_-_2108035
|
0.917
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chrX_-_128485134
|
0.884
|
NM_003069
NM_139035
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chr11_+_111288686
|
0.875
|
|
HSPB2-C11ORF52
HSPB2
|
HSPB2-C11orf52 read-through transcript
heat shock 27kDa protein 2
|
|
chr7_+_129919157
|
0.869
|
NM_002402
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr7_+_129918408
|
0.843
|
NM_177525
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr19_-_54835162
|
0.839
|
NM_006270
|
RRAS
|
related RAS viral (r-ras) oncogene homolog
|
|
chrX_-_128485121
|
0.818
|
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chrX_-_128485062
|
0.798
|
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chr9_-_112840064
|
0.789
|
NM_001401
NM_057159
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr11_+_19328846
|
0.785
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chr3_-_124893875
|
0.781
|
|
MYLK
|
myosin light chain kinase
|
|
chr4_-_102486975
|
0.762
|
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme
|
|
chr9_+_123101701
|
0.750
|
NM_000177
|
GSN
|
gelsolin
|
|
chr3_-_55490401
|
0.748
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr10_-_95350949
|
0.732
|
NM_006744
|
RBP4
|
retinol binding protein 4, plasma
|
|
chr2_+_235525355
|
0.720
|
NM_014521
|
SH3BP4
|
SH3-domain binding protein 4
|
|
chr4_-_102487356
|
0.710
|
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme
|
|
chr10_-_62373828
|
0.706
|
NM_014836
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chr9_+_123101926
|
0.682
|
|
GSN
|
gelsolin
|
|
chr12_+_6179736
|
0.674
|
NM_001769
|
CD9
|
CD9 molecule
|
|
chr21_-_27139143
|
0.669
|
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr16_+_54070302
|
0.666
|
NM_004530
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr11_+_44074322
|
0.630
|
NM_000401
|
EXT2
|
exostosin 2
|
|
chr6_-_128883125
|
0.627
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr21_-_27139563
|
0.627
|
NM_006988
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chrX_+_51653348
|
0.621
|
NM_001005333
NM_006986
|
MAGED1
|
melanoma antigen family D, 1
|
|
chrX_+_133993984
|
0.620
|
NM_001078171
|
FAM127A
|
family with sequence similarity 127, member A
|
|
chr9_+_111850698
|
0.615
|
NM_001004065
NM_001198656
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr15_-_27901828
|
0.615
|
NM_003257
NM_175610
|
TJP1
|
tight junction protein 1 (zona occludens 1)
|
|
chr22_+_36531178
|
0.606
|
|
H1F0
|
H1 histone family, member 0
|
|
chr1_-_85703321
|
0.604
|
NM_012137
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr3_-_151171576
|
0.602
|
|
PFN2
|
profilin 2
|
|
chr22_+_36531246
|
0.602
|
|
H1F0
|
H1 histone family, member 0
|
|
chr21_-_26465005
|
0.593
|
NM_000484
NM_001136129
NM_001136130
NM_201413
NM_201414
|
APP
|
amyloid beta (A4) precursor protein
|
|
chr16_+_55230078
|
0.586
|
NM_005946
|
MT1A
|
metallothionein 1A
|
|
chr14_-_50480857
|
0.570
|
NM_001163940
NM_002863
|
PYGL
|
phosphorylase, glycogen, liver
|
|
chrX_+_51944658
|
0.568
|
NM_001098800
NM_030801
NM_177535
NM_177537
|
MAGED4
MAGED4B
|
melanoma antigen family D, 4
melanoma antigen family D, 4B
|
|
chr12_-_105165805
|
0.565
|
NM_006825
|
CKAP4
|
cytoskeleton-associated protein 4
|
|
chr2_+_217206327
|
0.564
|
NM_000597
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa
|
|
chr2_+_217206450
|
0.554
|
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa
|
|
chr2_-_175578156
|
0.547
|
NM_001025201
NM_001822
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr12_+_51777684
|
0.544
|
NM_002178
|
IGFBP6
|
insulin-like growth factor binding protein 6
|
|
chr1_+_201862552
|
0.534
|
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4
|
|
chr11_-_2115086
|
0.530
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr21_+_41461924
|
0.525
|
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chr1_+_181258952
|
0.522
|
NM_002293
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2)
|
|
chr12_+_51777715
|
0.519
|
|
IGFBP6
|
insulin-like growth factor binding protein 6
|
|
chr6_+_142664490
|
0.508
|
NM_001032394
NM_001032395
NM_020455
NM_198569
|
GPR126
|
G protein-coupled receptor 126
|
|
chr4_+_166519392
|
0.507
|
NM_001873
|
CPE
|
carboxypeptidase E
|
|
chr10_+_124124196
|
0.506
|
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr3_-_151171190
|
0.503
|
NM_002628
NM_053024
|
PFN2
|
profilin 2
|
|
chr15_+_22772545
|
0.499
|
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N
|
|
chr17_+_58058783
|
0.499
|
|
MRC2
|
mannose receptor, C type 2
|
|
chrX_-_48818585
|
0.485
|
NM_007213
|
PRAF2
|
PRA1 domain family, member 2
|
|
chr5_+_71438796
|
0.480
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr6_+_151603346
|
0.468
|
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr5_-_168204430
|
0.467
|
|
|
|
|
chr2_+_219991342
|
0.466
|
NM_001927
|
DES
|
desmin
|
|
chr8_+_22492586
|
0.466
|
|
PDLIM2
|
PDZ and LIM domain 2 (mystique)
|
|
chr11_+_131285747
|
0.459
|
NM_001144058
NM_001144059
NM_016522
|
NTM
|
neurotrimin
|
|
chr20_-_17610828
|
0.457
|
NM_001042576
NM_004587
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr7_-_149669622
|
0.456
|
NM_002889
|
RARRES2
|
retinoic acid receptor responder (tazarotene induced) 2
|
|
chr16_+_30952382
|
0.452
|
NM_004604
|
STX4
|
syntaxin 4
|
|
chr1_-_151804929
|
0.444
|
NM_005978
|
S100A2
|
S100 calcium binding protein A2
|
|
chr8_-_67687980
|
0.440
|
NM_001080416
NM_001144755
|
MYBL1
LOC645895
|
v-myb myeloblastosis viral oncogene homolog (avian)-like 1
hypothetical LOC645895
|
|
chr20_-_61728774
|
0.439
|
NM_012384
|
GMEB2
|
glucocorticoid modulatory element binding protein 2
|
|
chr7_-_150576660
|
0.438
|
NM_001003801
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3
|
|
chr20_+_337346
|
0.433
|
|
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1
|
|
chr3_-_31997738
|
0.431
|
|
OSBPL10
|
oxysterol binding protein-like 10
|
|
chr14_+_64076916
|
0.431
|
NM_021979
|
HSPA2
|
heat shock 70kDa protein 2
|
|
chr21_-_26464915
|
0.425
|
|
APP
|
amyloid beta (A4) precursor protein
|
|
chrX_-_13866451
|
0.425
|
|
GPM6B
|
glycoprotein M6B
|
|
chr8_-_13035102
|
0.422
|
NM_006094
|
DLC1
|
deleted in liver cancer 1
|
|
chr7_+_116099648
|
0.419
|
NM_000245
NM_001127500
|
MET
|
met proto-oncogene (hepatocyte growth factor receptor)
|
|
chr2_-_26058941
|
0.409
|
NM_002254
|
KIF3C
|
kinesin family member 3C
|
|
chrX_+_54852217
|
0.409
|
NM_201222
|
MAGED2
|
melanoma antigen family D, 2
|
|
chr14_+_64077171
|
0.408
|
|
HSPA2
|
heat shock 70kDa protein 2
|
|
chr16_+_30952350
|
0.408
|
|
STX4
|
syntaxin 4
|
|
chr2_-_26058831
|
0.407
|
|
KIF3C
|
kinesin family member 3C
|
|
chr7_-_150576957
|
0.405
|
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3
|
|
chr6_+_151603201
|
0.403
|
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr2_+_8739563
|
0.403
|
NM_002166
|
ID2
|
inhibitor of DNA binding 2, dominant negative helix-loop-helix protein
|
|
chr1_+_181259317
|
0.401
|
|
|
|
|
chr1_+_201862548
|
0.401
|
NM_001001396
NM_001684
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4
|
|
chrX_-_119487096
|
0.395
|
|
LAMP2
|
lysosomal-associated membrane protein 2
|
|
chr1_-_85703198
|
0.392
|
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr16_+_30951915
|
0.392
|
|
STX4
|
syntaxin 4
|
|
chr20_+_25176698
|
0.382
|
NM_002862
|
PYGB
|
phosphorylase, glycogen; brain
|
|
chr9_-_35679799
|
0.380
|
|
TPM2
|
tropomyosin 2 (beta)
|
|
chr11_-_27697768
|
0.379
|
NM_001143807
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr2_+_202607554
|
0.379
|
NM_003507
|
FZD7
|
frizzled homolog 7 (Drosophila)
|
|
chr20_+_61798446
|
0.378
|
NM_003823
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy
|
|
chr6_+_148705817
|
0.375
|
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr9_-_112840801
|
0.374
|
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr14_-_104515738
|
0.374
|
NM_138420
|
AHNAK2
|
AHNAK nucleoprotein 2
|
|
chr4_+_74493218
|
0.373
|
|
ALB
|
albumin
|
|
chr8_+_31616809
|
0.372
|
NM_013962
|
NRG1
|
neuregulin 1
|
|
chrX_-_37591760
|
0.371
|
NM_006520
|
DYNLT3
|
dynein, light chain, Tctex-type 3
|
|
chr13_-_109236897
|
0.371
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr1_-_27802947
|
0.371
|
|
AHDC1
|
AT hook, DNA binding motif, containing 1
|
|
chrX_-_119487164
|
0.369
|
|
LAMP2
|
lysosomal-associated membrane protein 2
|
|
chr17_-_6958695
|
0.367
|
|
ASGR2
|
asialoglycoprotein receptor 2
|
|
chr9_-_73573121
|
0.365
|
NM_001135820
NM_013390
|
TMEM2
|
transmembrane protein 2
|
|
chr6_-_35572694
|
0.364
|
NM_003214
|
TEAD3
|
TEA domain family member 3
|
|
chr7_+_79602023
|
0.362
|
NM_002069
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1
|
|
chr4_-_54625463
|
0.361
|
|
CHIC2
|
cysteine-rich hydrophobic domain 2
|
|
chr20_+_48240705
|
0.361
|
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta
|
|
chrX_-_13866565
|
0.361
|
NM_001001994
|
GPM6B
|
glycoprotein M6B
|
|
chr9_-_35679902
|
0.359
|
NM_003289
NM_213674
|
TPM2
|
tropomyosin 2 (beta)
|
|
chr5_+_131621287
|
0.356
|
|
PDLIM4
|
PDZ and LIM domain 4
|
|
chr4_+_74493331
|
0.356
|
|
ALB
|
albumin
|
|
chrX_-_37591703
|
0.352
|
|
DYNLT3
|
dynein, light chain, Tctex-type 3
|
|
chr10_+_170423
|
0.346
|
NM_001161482
NM_006624
NM_212479
|
ZMYND11
|
zinc finger, MYND-type containing 11
|
|
chr18_+_54013575
|
0.344
|
NM_001144970
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr20_-_36191623
|
0.338
|
|
TGM2
|
transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase)
|
|
chr6_+_148705646
|
0.337
|
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr4_-_102487649
|
0.336
|
NM_000944
NM_001130691
NM_001130692
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme
|
|
chr18_+_7557313
|
0.336
|
NM_001105244
NM_002845
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chr2_-_220144427
|
0.334
|
|
OBSL1
|
obscurin-like 1
|
|
chr4_+_74493266
|
0.332
|
|
ALB
|
albumin
|
|
chr9_-_122516399
|
0.332
|
NM_001080497
|
MEGF9
|
multiple EGF-like-domains 9
|
|
chr20_+_48240772
|
0.332
|
NM_005194
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta
|
|
chr9_-_87904844
|
0.331
|
NM_177937
|
GOLM1
|
golgi membrane protein 1
|
|
chr16_-_66827449
|
0.331
|
NM_024939
|
ESRP2
|
epithelial splicing regulatory protein 2
|
|
chrX_+_17665483
|
0.323
|
NM_001037535
NM_001037536
NM_001037540
NM_006746
|
SCML1
|
sex comb on midleg-like 1 (Drosophila)
|
|
chr11_-_70350292
|
0.321
|
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr19_+_8384181
|
0.321
|
NM_001005415
NM_001005416
NM_016496
|
MARCH2
|
membrane-associated ring finger (C3HC4) 2
|
|
chr19_-_12747421
|
0.319
|
NM_001100176
NM_013312
|
HOOK2
|
hook homolog 2 (Drosophila)
|
|
chr5_+_86599350
|
0.318
|
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1
|
|
chr16_-_65343012
|
0.318
|
NM_006141
|
DYNC1LI2
|
dynein, cytoplasmic 1, light intermediate chain 2
|
|
chr3_-_95175588
|
0.317
|
NM_000313
|
PROS1
|
protein S (alpha)
|
|
chr1_-_93919789
|
0.316
|
NM_003567
|
BCAR3
|
breast cancer anti-estrogen resistance 3
|
|
chr10_+_124124083
|
0.315
|
NM_001001974
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr2_+_62787534
|
0.315
|
NM_001142614
|
EHBP1
|
EH domain binding protein 1
|
|
chr6_+_1335067
|
0.315
|
NM_001452
|
FOXF2
|
forkhead box F2
|
|
chr11_-_61415429
|
0.314
|
NM_021727
|
FADS3
|
fatty acid desaturase 3
|
|
chr12_+_12935222
|
0.313
|
NM_003979
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chrX_-_108863109
|
0.313
|
|
ACSL4
|
acyl-CoA synthetase long-chain family member 4
|
|
chr5_-_141684759
|
0.313
|
NM_001127496
NM_030964
|
SPRY4
|
sprouty homolog 4 (Drosophila)
|
|
chr11_-_35503720
|
0.311
|
NM_001001991
NM_015430
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr7_+_94130673
|
0.310
|
|
PEG10
|
paternally expressed 10
|
|
chr20_+_43874735
|
0.307
|
|
UBE2C
|
ubiquitin-conjugating enzyme E2C
|
|
chr2_+_20510312
|
0.306
|
NM_004040
|
RHOB
|
ras homolog gene family, member B
|
|
chr3_-_187025501
|
0.303
|
NM_001007225
NM_006548
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr4_-_54625223
|
0.303
|
|
CHIC2
|
cysteine-rich hydrophobic domain 2
|
|
chr1_+_43837096
|
0.302
|
|
PTPRF
|
protein tyrosine phosphatase, receptor type, F
|
|
chr8_-_145097031
|
0.301
|
NM_201380
|
PLEC
|
plectin
|
|
chr12_+_77782719
|
0.300
|
|
SYT1
|
synaptotagmin I
|
|
chr12_+_12935619
|
0.298
|
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr22_-_49063088
|
0.297
|
|
PLXNB2
|
plexin B2
|
|
chrX_+_101792949
|
0.296
|
NM_001099410
NM_001099411
NM_014710
NM_001184727
|
GPRASP1
|
G protein-coupled receptor associated sorting protein 1
|
|
chr17_-_15109315
|
0.295
|
NM_000304
|
PMP22
|
peripheral myelin protein 22
|
|
chr19_+_627346
|
0.293
|
NM_005860
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein)
|
|
chr5_+_131621276
|
0.291
|
|
PDLIM4
|
PDZ and LIM domain 4
|
|
chr19_-_12747263
|
0.288
|
|
HOOK2
|
hook homolog 2 (Drosophila)
|
|
chr7_+_134114957
|
0.287
|
|
CALD1
|
caldesmon 1
|
|
chr12_+_109536214
|
0.284
|
NM_001082537
NM_001082538
NM_001173976
NM_024549
NM_001173975
|
TCTN1
|
tectonic family member 1
|
|
chrX_+_70359780
|
0.284
|
NM_000166
|
GJB1
|
gap junction protein, beta 1, 32kDa
|
|
chr9_-_130912728
|
0.282
|
NM_000755
|
CRAT
|
carnitine O-acetyltransferase
|
|
chr14_+_105012106
|
0.282
|
NM_001312
|
CRIP2
|
cysteine-rich protein 2
|
|
chr4_+_74493174
|
0.280
|
|
ALB
|
albumin
|
|
chr11_+_113815317
|
0.280
|
NM_015523
|
REXO2
|
REX2, RNA exonuclease 2 homolog (S. cerevisiae)
|
|
chr2_-_20075889
|
0.280
|
|
MATN3
|
matrilin 3
|
|
chr12_-_6668880
|
0.280
|
NM_001039917
NM_001039919
NM_001039920
NM_001135734
|
ZNF384
|
zinc finger protein 384
|
|
chr2_-_20288647
|
0.279
|
NM_001006946
|
SDC1
|
syndecan 1
|
|
chr18_+_19706979
|
0.278
|
NM_000227
NM_001127718
|
LAMA3
|
laminin, alpha 3
|
|
chr12_+_2870255
|
0.277
|
NM_001160408
NM_003324
|
TULP3
|
tubby like protein 3
|
|
chr17_-_5344898
|
0.276
|
NM_001162371
|
LOC728392
|
hypothetical protein LOC728392
|
|
chr19_-_56147971
|
0.275
|
NM_001077491
NM_001077492
NM_012427
|
KLK5
|
kallikrein-related peptidase 5
|
|
chrX_+_102518764
|
0.275
|
NM_014380
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr8_+_9450849
|
0.274
|
NM_003747
|
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase
|
|
chr20_-_22512893
|
0.272
|
|
FOXA2
|
forkhead box A2
|
|
chr12_+_77782648
|
0.269
|
|
SYT1
|
synaptotagmin I
|
|
chr17_-_1342700
|
0.269
|
NM_001080779
|
MYO1C
|
myosin IC
|
|
chr12_-_48902692
|
0.267
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr14_+_100362236
|
0.266
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr20_-_17610704
|
0.266
|
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr5_+_131621230
|
0.266
|
NM_001131027
NM_003687
|
PDLIM4
|
PDZ and LIM domain 4
|
|
chr20_+_43874654
|
0.265
|
NM_007019
NM_181799
NM_181800
NM_181802
NM_181803
|
UBE2C
|
ubiquitin-conjugating enzyme E2C
|
|
chr19_-_10166536
|
0.265
|
|
DNMT1
|
DNA (cytosine-5-)-methyltransferase 1
|
|
chr20_+_43875078
|
0.263
|
NM_181801
|
UBE2C
|
ubiquitin-conjugating enzyme E2C
|
|
chr21_-_26464931
|
0.262
|
|
APP
|
amyloid beta (A4) precursor protein
|